Loading Data via Zarr Endpoints¶
Zarr Format Benefits:
- Scalable, flexible
- Easy to access via HTTP/HTTPS in cloud storage
But:
- Majority of datasets in HDF5 (netCDF4)
- HDF5/netCDF4 hard to access via HTTP/HTTPS in cloud storage
Freva Solution:
- REST API streams any file format as Zarr
- Zarr protocol endpoints accessible via any Zarr library
Workflow¶
- Search netCDF4 datasets using Freva-REST API
- Access data through Zarr endpoints
Let's define the search parameters for the Freva-REST API and import what we need
search_params = {"dataset": "cmip6-fs", "project": "cmip6"} # Define our search parameters
url = "http://localhost:7777" # URL of our test server.
from getpass import getpass
import requests
from tempfile import NamedTemporaryFile
import xarray as xr
If we normally search for data we will get the locations of the netCDF files on the hard-drive:
list(requests.get(
f"{url}/api/databrowser/data_search/freva/file",
params=search_params,
stream=True
).iter_lines(decode_unicode=True))
['/home/wilfred/workspace/freva-nextgen/freva-rest/src/databrowser_api/mock/data/model/global/cmip6/CMIP6/CMIP/MPI-M/MPI-ESM1-2-LR/amip/r2i1p1f1/Amon/ua/gn/v20190815/ua_mon_MPI-ESM1-2-LR_amip_r2i1p1f1_gn_197901-199812.nc', '/home/wilfred/workspace/freva-nextgen/freva-rest/src/databrowser_api/mock/data/model/global/cmip6/CMIP6/CMIP/CSIRO-ARCCSS/ACCESS-CM2/amip/r1i1p1f1/Amon/ua/gn/v20201108/ua_Amon_ACCESS-CM2_amip_r1i1p1f1_gn_197901-201412.nc']
What if the data location is not directly accessible, because it's stored somewhere else, like on tape?
- We can use the
loadendpoint to stream stream the data as Zarr data.
Caveat: Because the data can be accessed from anywhere once it is made available via zarr we need to create an access token:
auth = requests.post(
f"{url}/api/auth/v2/token",
data={"username": "janedoe", "password":getpass("Password: ")}
).json()
"Order" the zarr datasets.¶
With this access token we can generate zarr enpoints to stream the data from anywhere, to do so we simply search for the datasets again:
res = requests.get(
f"{url}/api/databrowser/load/freva",
params=search_params,
headers={
"Authorization": f"Bearer {auth['access_token']}"
},
stream=True
)
This will search for data and for every found entry create a zarr endpoint that can be loaded:
zarr_files = list(res.iter_lines(decode_unicode=True))
zarr_files
['http://localhost:7777/api/freva-data-portal/zarr/dcb608a0-9d77-5045-b656-f21dfb5e9acf.zarr', 'http://localhost:7777/api/freva-data-portal/zarr/f56264e3-d713-5c27-bc4e-c97f15b5fe86.zarr']
Open the zarr datasets¶
Let's load the data with xarray and zarr:
dset = xr.open_dataset(
zarr_files[0],
engine="zarr",
chunks="auto",
storage_options={"headers": {"Authorization": f"Bearer {auth['access_token']}"}}
)
dset
<xarray.Dataset>
Dimensions: (lat: 27, bnds: 2, lon: 43, plev: 19, time: 11)
Coordinates:
* lat (lat) float64 0.9326 2.798 4.663 6.528 ... 43.83 45.7 47.56 49.43
* lon (lon) float64 101.2 103.1 105.0 106.9 ... 174.4 176.2 178.1 180.0
* plev (plev) float64 1e+05 9.25e+04 8.5e+04 7e+04 ... 1e+03 500.0 100.0
* time (time) datetime64[ns] 1979-01-16T12:00:00 ... 1979-11-16
Dimensions without coordinates: bnds
Data variables:
lat_bnds (lat, bnds) float64 dask.array<chunksize=(27, 2), meta=np.ndarray>
lon_bnds (lon, bnds) float64 dask.array<chunksize=(43, 2), meta=np.ndarray>
time_bnds (time, bnds) datetime64[ns] dask.array<chunksize=(11, 2), meta=np.ndarray>
ua (time, plev, lat, lon) float32 dask.array<chunksize=(11, 19, 27, 43), meta=np.ndarray>
Attributes: (12/47)
CDI: Climate Data Interface version 2.0.6 (https://mpim...
source: MPI-ESM1.2-LR (2017): \naerosol: none, prescribed ...
institution: Max Planck Institute for Meteorology, Hamburg 2014...
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: no parent
... ...
variable_id: ua
variant_label: r2i1p1f1
license: CMIP6 model data produced by MPI-M is licensed und...
cmor_version: 3.5.0
tracking_id: hdl:21.14100/0898c2ad-5382-4d0c-8adb-2ca96387fb54
CDO: Climate Data Operators version 2.0.6 (https://mpim...- lat: 27
- bnds: 2
- lon: 43
- plev: 19
- time: 11
- lat(lat)float640.9326 2.798 4.663 ... 47.56 49.43
- standard_name :
- latitude
- long_name :
- Latitude
- units :
- degrees_north
- axis :
- Y
- bounds :
- lat_bnds
array([ 0.93263 , 2.79789 , 4.66315 , 6.528409, 8.393669, 10.258928, 12.124187, 13.989446, 15.854704, 17.719962, 19.585219, 21.450475, 23.315731, 25.180986, 27.046239, 28.911492, 30.776744, 32.641994, 34.507243, 36.372491, 38.237736, 40.102979, 41.96822 , 43.833459, 45.698694, 47.563926, 49.429154]) - lon(lon)float64101.2 103.1 105.0 ... 178.1 180.0
- standard_name :
- longitude
- long_name :
- Longitude
- units :
- degrees_east
- axis :
- X
- bounds :
- lon_bnds
array([101.25 , 103.125, 105. , 106.875, 108.75 , 110.625, 112.5 , 114.375, 116.25 , 118.125, 120. , 121.875, 123.75 , 125.625, 127.5 , 129.375, 131.25 , 133.125, 135. , 136.875, 138.75 , 140.625, 142.5 , 144.375, 146.25 , 148.125, 150. , 151.875, 153.75 , 155.625, 157.5 , 159.375, 161.25 , 163.125, 165. , 166.875, 168.75 , 170.625, 172.5 , 174.375, 176.25 , 178.125, 180. ]) - plev(plev)float641e+05 9.25e+04 ... 500.0 100.0
- standard_name :
- air_pressure
- long_name :
- pressure
- units :
- Pa
- positive :
- down
- axis :
- Z
array([100000., 92500., 85000., 70000., 60000., 50000., 40000., 30000., 25000., 20000., 15000., 10000., 7000., 5000., 3000., 2000., 1000., 500., 100.]) - time(time)datetime64[ns]1979-01-16T12:00:00 ... 1979-11-16
- standard_name :
- time
- long_name :
- time
- bounds :
- time_bnds
- axis :
- T
array(['1979-01-16T12:00:00.000000000', '1979-02-15T00:00:00.000000000', '1979-03-16T12:00:00.000000000', '1979-04-16T00:00:00.000000000', '1979-05-16T12:00:00.000000000', '1979-06-16T00:00:00.000000000', '1979-07-16T12:00:00.000000000', '1979-08-16T12:00:00.000000000', '1979-09-16T00:00:00.000000000', '1979-10-16T12:00:00.000000000', '1979-11-16T00:00:00.000000000'], dtype='datetime64[ns]')
- lat_bnds(lat, bnds)float64dask.array<chunksize=(27, 2), meta=np.ndarray>
Array Chunk Bytes 432 B 432 B Shape (27, 2) (27, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - lon_bnds(lon, bnds)float64dask.array<chunksize=(43, 2), meta=np.ndarray>
Array Chunk Bytes 688 B 688 B Shape (43, 2) (43, 2) Dask graph 1 chunks in 2 graph layers Data type float64 numpy.ndarray - time_bnds(time, bnds)datetime64[ns]dask.array<chunksize=(11, 2), meta=np.ndarray>
Array Chunk Bytes 176 B 176 B Shape (11, 2) (11, 2) Dask graph 1 chunks in 2 graph layers Data type datetime64[ns] numpy.ndarray - ua(time, plev, lat, lon)float32dask.array<chunksize=(11, 19, 27, 43), meta=np.ndarray>
- standard_name :
- eastward_wind
- long_name :
- Eastward Wind
- units :
- m s-1
- CDI_grid_type :
- gaussian
- CDI_grid_num_LPE :
- 48
- comment :
- Zonal wind (positive in a eastward direction).
- cell_methods :
- time: mean
- cell_measures :
- area: areacella
- history :
- 2020-06-05T18:27:23Z altered by CMOR: Reordered dimensions, original order: time lat lon plev. 2020-06-05T18:27:23Z altered by CMOR: replaced missing value flag (-9e+33) and corresponding data with standard missing value (1e+20). 2020-06-05T18:27:23Z altered by CMOR: Inverted axis: lat.
Array Chunk Bytes 0.93 MiB 0.93 MiB Shape (11, 19, 27, 43) (11, 19, 27, 43) Dask graph 1 chunks in 2 graph layers Data type float32 numpy.ndarray
- latPandasIndex
PandasIndex(Index([ 0.932629967837991, 2.797889876956741, 4.663149706177884, 6.5284094014799905, 8.393668907692383, 10.258928168006376, 12.124187123455766, 13.989445712356673, 15.854703869694873, 17.719961526447428, 19.58521860882233, 21.450475037398185, 23.31573072614093, 25.180985581270594, 27.04623949994481, 28.91149236871774, 30.77674406172325, 32.64199443851768, 34.50724334150103, 36.37249059281224, 38.23773599056483, 40.1029793042494, 41.96822026907538, 43.83345857895126, 45.698693877701785, 47.56392574797867, 49.42915369712305], dtype='float64', name='lat')) - lonPandasIndex
PandasIndex(Index([ 101.25, 103.125, 105.0, 106.875, 108.75, 110.625, 112.5, 114.375, 116.25, 118.125, 120.0, 121.875, 123.75, 125.625, 127.5, 129.375, 131.25, 133.125, 135.0, 136.875, 138.75, 140.625, 142.5, 144.375, 146.25, 148.125, 150.0, 151.875, 153.75, 155.625, 157.5, 159.375, 161.25, 163.125, 165.0, 166.875, 168.75, 170.625, 172.5, 174.375, 176.25, 178.125, 180.0], dtype='float64', name='lon')) - plevPandasIndex
PandasIndex(Index([100000.0, 92500.0, 85000.0, 70000.0, 60000.0, 50000.0, 40000.0, 30000.0, 25000.0, 20000.0, 15000.0, 10000.0, 7000.0, 5000.0, 3000.0, 2000.0, 1000.0, 500.0, 100.0], dtype='float64', name='plev')) - timePandasIndex
PandasIndex(DatetimeIndex(['1979-01-16 12:00:00', '1979-02-15 00:00:00', '1979-03-16 12:00:00', '1979-04-16 00:00:00', '1979-05-16 12:00:00', '1979-06-16 00:00:00', '1979-07-16 12:00:00', '1979-08-16 12:00:00', '1979-09-16 00:00:00', '1979-10-16 12:00:00', '1979-11-16 00:00:00'], dtype='datetime64[ns]', name='time', freq=None))
- CDI :
- Climate Data Interface version 2.0.6 (https://mpimet.mpg.de/cdi)
- source :
- MPI-ESM1.2-LR (2017): aerosol: none, prescribed MACv2-SP atmos: ECHAM6.3 (spectral T63; 192 x 96 longitude/latitude; 47 levels; top level 0.01 hPa) atmosChem: none land: JSBACH3.20 landIce: none/prescribed ocean: MPIOM1.63 (bipolar GR1.5, approximately 1.5deg; 256 x 220 longitude/latitude; 40 levels; top grid cell 0-12 m) ocnBgchem: HAMOCC6 seaIce: unnamed (thermodynamic (Semtner zero-layer) dynamic (Hibler 79) sea ice model)
- institution :
- Max Planck Institute for Meteorology, Hamburg 20146, Germany
- Conventions :
- CF-1.7 CMIP-6.2
- activity_id :
- CMIP
- branch_method :
- no parent
- contact :
- cmip6-mpi-esm@dkrz.de
- creation_date :
- 2020-06-05T18:27:23Z
- data_specs_version :
- 01.00.30
- experiment :
- AMIP
- experiment_id :
- amip
- external_variables :
- areacella
- forcing_index :
- 1
- frequency :
- mon
- further_info_url :
- https://furtherinfo.es-doc.org/CMIP6.MPI-M.MPI-ESM1-2-LR.amip.none.r2i1p1f1
- grid :
- gn
- grid_label :
- gn
- history :
- Thu Sep 29 13:40:53 2022: cdo sellonlatbox,100,180,0,50 ua_mon_MPI-ESM1-2-LR_amip_r2i1p1f1_gn_197901-199812.nc tmp.nc 2020-06-05T18:27:23Z ; CMOR rewrote data to be consistent with CMIP6, CF-1.7 CMIP-6.2 and CF standards.
- initialization_index :
- 1
- institution_id :
- MPI-M
- mip_era :
- CMIP6
- nominal_resolution :
- 250 km
- parent_activity_id :
- no parent
- parent_experiment_id :
- no parent
- parent_mip_era :
- no parent
- parent_source_id :
- no parent
- parent_time_units :
- no parent
- parent_variant_label :
- no parent
- physics_index :
- 1
- product :
- model-output
- project_id :
- CMIP6
- realization_index :
- 2
- realm :
- atmos
- references :
- MPI-ESM: Mauritsen, T. et al. (2019), Developments in the MPI‐M Earth System Model version 1.2 (MPI‐ESM1.2) and Its Response to Increasing CO2, J. Adv. Model. Earth Syst.,11, 998-1038, doi:10.1029/2018MS001400, Mueller, W.A. et al. (2018): A high‐resolution version of the Max Planck Institute Earth System Model MPI‐ESM1.2‐HR. J. Adv. Model. EarthSyst.,10,1383–1413, doi:10.1029/2017MS001217
- source_id :
- MPI-ESM1-2-LR
- source_type :
- AGCM
- sub_experiment :
- none
- sub_experiment_id :
- none
- table_id :
- Amon
- table_info :
- Creation Date:(09 May 2019) MD5:dff4d5e7b285678699ef52ab1a3cca43
- title :
- MPI-ESM1-2-LR output prepared for CMIP6
- variable_id :
- ua
- variant_label :
- r2i1p1f1
- license :
- CMIP6 model data produced by MPI-M is licensed under a Creative Commons Attribution ShareAlike 4.0 International License (https://creativecommons.org/licenses). Consult https://pcmdi.llnl.gov/CMIP6/TermsOfUse for terms of use governing CMIP6 output, including citation requirements and proper acknowledgment. Further information about this data, including some limitations, can be found via the further_info_url (recorded as a global attribute in this file) and. The data producers and data providers make no warranty, either express or implied, including, but not limited to, warranties of merchantability and fitness for a particular purpose. All liabilities arising from the supply of the information (including any liability arising in negligence) are excluded to the fullest extent permitted by law.
- cmor_version :
- 3.5.0
- tracking_id :
- hdl:21.14100/0898c2ad-5382-4d0c-8adb-2ca96387fb54
- CDO :
- Climate Data Operators version 2.0.6 (https://mpimet.mpg.de/cdo)
We do have a xarray dataset, meaning we can just proceed with our analysis:
dset["ua"].mean(dim=("lon", "lat")).plot(x="time", yincrease=False)
<matplotlib.collections.QuadMesh at 0x7f7b7ecefad0>
Creating intake catalogues¶
Intake can conviniently aggregate data. Instead creating individual lists of files we can create an intake catalogue with the zarr end points that helps us to aggregate the data later.
To create an intake catalogue instead of a list of files we simply have to add the catalogue-type:intake search parameter:
import intake
search_params["catalogue-type"] = "intake"
res = requests.get(
f"{url}/api/databrowser/load/freva",
params=search_params,
headers={
"Authorization": f"Bearer {auth['access_token']}"
},
stream=True
)
with NamedTemporaryFile(suffix=".json") as temp_f:
with open(temp_f.name, "w") as stream:
stream.write(res.text)
cat = intake.open_esm_datastore(temp_f.name)
cat.df
| uri | project | product | institute | model | experiment | time_frequency | realm | variable | ensemble | cmor_table | fs_type | grid_label | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | http://localhost:7777/api/freva-data-portal/za... | CMIP6 | CMIP | MPI-M | MPI-ESM1-2-LR | amip | mon | atmos | ua | r2i1p1f1 | Amon | posix | gn |
| 1 | http://localhost:7777/api/freva-data-portal/za... | CMIP6 | CMIP | CSIRO-ARCCSS | ACCESS-CM2 | amip | mon | atmos | ua | r1i1p1f1 | Amon | posix | gn |
Using the freva client libray¶
Rest requests can be confusing for many users. The new freva_client library is here to help
from freva_client import authenticate, databrowser
data_query = databrowser(dataset="cmip6-fs", host="localhost:7777", stream_zarr=True)
token = authenticate(username="janedoe", host="localhost:7777")
files = list(data_query)
files
['http://localhost:7777/api/freva-data-portal/zarr/dcb608a0-9d77-5045-b656-f21dfb5e9acf.zarr', 'http://localhost:7777/api/freva-data-portal/zarr/f56264e3-d713-5c27-bc4e-c97f15b5fe86.zarr']
We can also use the freva client library to directly create an intake catalogue:
cat = data_query.intake_catalogue()
cat.df
| uri | project | product | institute | model | experiment | time_frequency | realm | variable | ensemble | cmor_table | fs_type | grid_label | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | http://localhost:7777/api/freva-data-portal/za... | CMIP6 | CMIP | MPI-M | MPI-ESM1-2-LR | amip | mon | atmos | ua | r2i1p1f1 | Amon | posix | gn |
| 1 | http://localhost:7777/api/freva-data-portal/za... | CMIP6 | CMIP | CSIRO-ARCCSS | ACCESS-CM2 | amip | mon | atmos | ua | r1i1p1f1 | Amon | posix | gn |
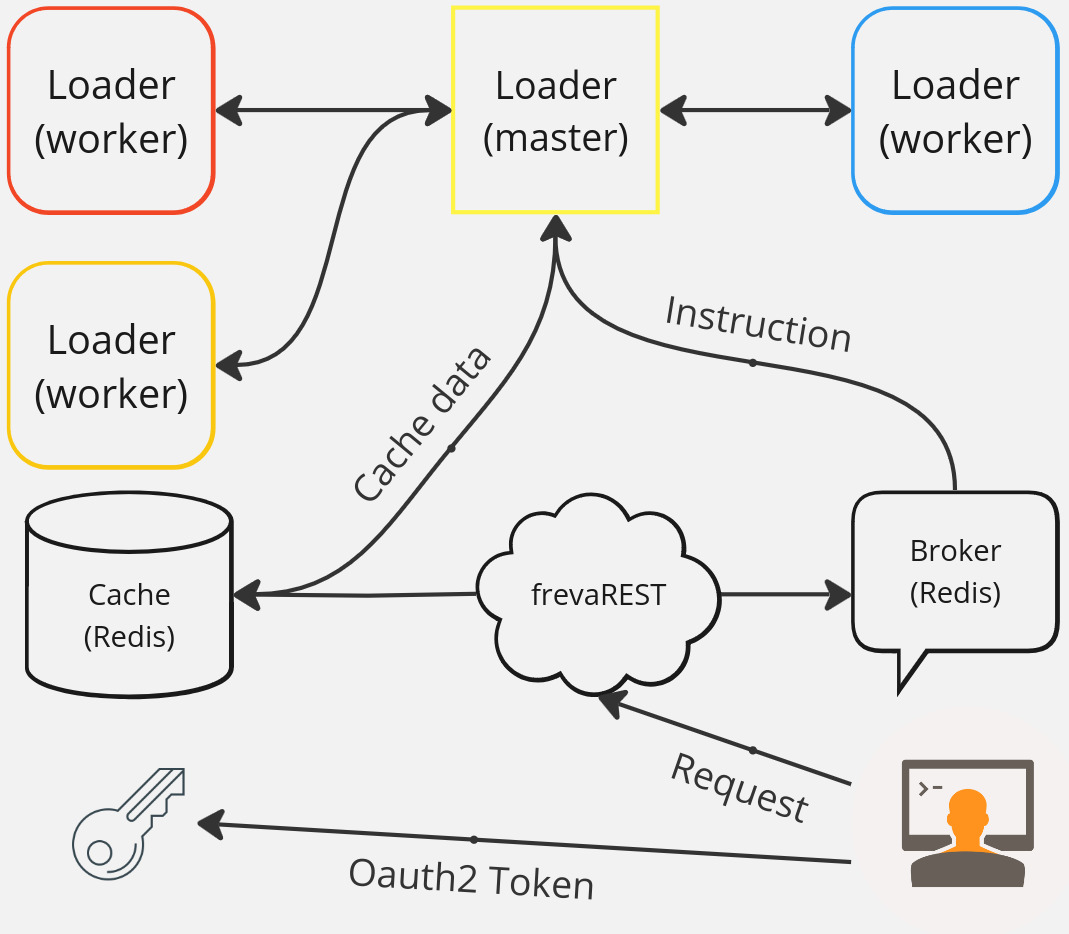
How does it work?¶
What's next?¶
- Add json payload to
loadendpoint that allows the users to pre-precess data. For example select a region by uploading a geojson shape file. - Implement a backend handle to open tape archives