Freva python module#
The following section gives an overview over the usage of the Freva python module. This section assumes that you know how to get to access to the python environment that has Freva installed. If this is not the case please contact one of your Freva admins or the Frequently Asked Questions section for help.
Searching for data#
To query data databrowser and search for data you have three different options. You can the to following methods
freva.databrowser(): The main method for searching data is thefreva.databrowser()method. The data browser method lets you search for data files or uris (Uniform Resource Identifier). Uris instead of file paths are useful because an uri indicates the storage system where the files are located.freva.facet_search(): This method lists all search categories (facets) and their values.freva.count_values(): You can count the occurrences of search results with this method.
Below you can find a more detailed documentation.
- freva.count_values(*, time: str = '', time_select: Literal['strict', 'flexible', 'file'] = 'flexible', multiversion: bool = False, facet: str | list[str] | None = None, **search_facets: str | list[str] | int) int | dict[str, dict[str, int]]#
Count the number of found objects in the databrowser.
- Parameters:
time (str, default: "") – Special search facet to refine/subset search results by time. This can be a string representation of a time range or a single time step. The time steps have to follow ISO-8601. Valid strings are
%Y-%m-%dT%H:%Mto%Y-%m-%dT%H:%Mfor time ranges and%Y-%m-%dT%H:%M. Note: You don’t have to give the full string format to subset time steps%Y,%Y-%metc are also valid.time_select (str, default: flexible) – Operator that specifies how the time period is selected. Choose from flexible (default), strict or file.
strictreturns only those files that have the entire time period covered. The time search2000 to 2012will not select files containing data from 2010 to 2020 with thestrictmethod.flexiblewill select those files asflexiblereturns those files that have either start or end period covered.filewill only return files where the entire time period is contained within one single file.multiversion (bool, default: False) – Select all versions and not just the latest version (default).
facet (Union[str, list[str]], default: None) – Count these these facets (attributes & values) instead of the number of total files. If None (default), the number of total files will be returned.
**search_facets (str) – The facets to be applied in the data search. If not given the whole dataset will be queried.
- Returns:
Number of found objects, if the facet key is/are given then the a dictionary with the number of objects for each search facet/key is given.
- Return type:
int, dict[str, int]
Example
Code
import freva num_files = freva.count_values(experiment="cmorph") print(num_files)
Results
24
Code
import freva print(freva.count_values(facet="*"))
Results
{'fs_type': {'posix': 39}, 'cmor_table': {'30min': 24, '3hr': 2, 'aday': 10, 'amon': 2, 'omon': 1}, 'grid_label': {'gn': 39}, 'realm': {'atmos': 38, 'ocean': 1}, 'institute': {'clmcom': 10, 'cpc': 24, 'csiro-arccss': 1, 'gerics': 2, 'mpi-m': 1, 'noaa': 1}, 'time_aggregation': {'mean': 39}, 'future': {}, 'variable': {'hc700': 1, 'pr': 26, 'tas': 10, 'ua': 2}, 'model': {'access-cm2': 1, 'cpc': 24, 'mpi-esm1-2-lr': 1, 'mpi-m-mpi-esm-lr-clmcom-cclm4-8-17-v1': 10, 'ncc-noresm1-m-gerics-remo2015-v1': 2, 'nodc': 1}, 'ensemble': {'r1i1p1': 37, 'r1i1p1f1': 1, 'r2i1p1f1': 1}, 'project': {'cmip6': 2, 'cordex': 12, 'observations': 24, 'reanalysis': 1}, 'future_id': {}, 'experiment': {'amip': 2, 'cmorph': 24, 'historical': 10, 'oc5': 1, 'rcp85': 2}, 'time_frequency': {'1day': 10, '30min': 24, '3hr': 2, 'mon': 3}, 'product': {'cmip': 2, 'eur-11': 12, 'grid': 24, 'reanalysis': 1}, 'bbox': {}}
- freva.databrowser(*, multiversion: bool = False, batch_size: int = 5000, uniq_key: Literal['file', 'uri'] = 'file', time: str = '', time_select: Literal['flexible', 'strict', 'file'] = 'flexible', **search_facets: str | list[str] | int) dict[str, dict[str, int]] | dict[str, list[str]] | Iterator[str] | int#
Find data in the system.
You can either search for files or data facets (variable, model, …) that are available. The query is of the form key=value. <value> might use *, ? as wildcards or any regular expression.
- Parameters:
**search_facets (Union[str, Path, in, list[str]]) – The facets to be applied in the data search. If not given the whole dataset will be queried.
time (str) – Special search facet to refine/subset search results by time. This can be a string representation of a time range or a single time step. The time steps have to follow ISO-8601. Valid strings are
%Y-%m-%dT%H:%Mto%Y-%m-%dT%H:%Mfor time ranges and%Y-%m-%dT%H:%M. Note: You don’t have to give the full string format to subset time steps%Y,%Y-%metc are also valid.time_select (str, default: flexible) – Operator that specifies how the time period is selected. Choose from flexible (default), strict or file.
strictreturns only those files that have the entire time period covered. The time search2000 to 2012will not select files containing data from 2010 to 2020 with thestrictmethod.flexiblewill select those files asflexiblereturns those files that have either start or end period covered.filewill only return files where the entire time period is contained within one single file.uniq_key (str, default: file) – Chose if the solr search query should return paths to files or uris, uris will have the file path along with protocol of the storage system. Uris can be useful if the the search query result should be used libraries like fsspec.
multiversion (bool, default: False) – Select all versions and not just the latest version (default).
batch_size (int, default: 5000) – Size of the search query.
- Returns:
If
all_facetsis False andfacetis None an iterator with results.- Return type:
Iterator
Example
Search for files in the system:
Code
import freva files = freva.databrowser(project='obs*', institute='cpc', time_frequency='??min', variable='pr') print(files) print(next(files)) for file in files: print(file) break
Results
<generator object SolrFindFiles._search at 0x7f71b6769e70> /home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022300-201609022330.nc /home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022200-201609022230.nc
Search for files between a two given time steps:
Code
import freva file_range = freva.databrowser(project="obs*", time="2016-09-02T22:15 to 2016-10") for file in file_range: print(file)
Results
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022300-201609022330.nc /home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022200-201609022230.nc
The default method for selecting time periods is
flexible, which means all files are selected that cover at least start or end date. Thestrictmethod implies that the entire search time period has to be covered by the files. Using thestrictmethod in the example above would only yield one file because the first file contains time steps prior to the start of the time period:Code
import freva file_range = freva.databrowser(project="obs*", time="2016-09-02T22:15 to 2016-10", time_select="strict") for file in file_range: print(file)
Results
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022300-201609022330.nc
In datasets with multiple versions only the latest version (i.e. highest version number) is returned by default. Querying a specific version from a multi versioned datasets requires the
multiversionflag in combination with theversionspecial facet:Code
import freva specific_version = list(freva.databrowser(project="reanalysis", version="v20200101", multiversion=True)) print(specific_version)
Results
['/home/runner/work/freva/freva/.docker/data/reanalysis/reanalysis/NOAA/NODC/OC5/mon/ocean/Omon/r1i1p1/v20200101/hc700/hc700_mon_NODC_OC5_r1i1p1_201201-201212.nc']
- freva.facet_search(*, time: str = '', time_select: Literal['strict', 'flexible', 'file'] = 'flexible', multiversion: bool = False, facet: str | list[str] | None = None, **search_facets: str | list[str] | int) dict[str, list[str]]#
Search for data attributes (facets) in the databrowser.
The method queries the databrowser for available search facets (keys) like model, experiment etc.
- Parameters:
time (str) – Special search facet to refine/subset search results by time. This can be a string representation of a time range or a single time step. The time steps have to follow ISO-8601. Valid strings are
%Y-%m-%dT%H:%Mto%Y-%m-%dT%H:%Mfor time ranges and%Y-%m-%dT%H:%M. Note: You don’t have to give the full string format to subset time steps%Y,%Y-%metc are also valid.time_select (str, default: flexible) – Operator that specifies how the time period is selected. Choose from flexible (default), strict or file.
strictreturns only those files that have the entire time period covered. The time search2000 to 2012will not select files containing data from 2010 to 2020 with thestrictmethod.flexiblewill select those files asflexiblereturns those files that have either start or end period covered.filewill only return files where the entire time period is contained within one single file.facet (Union[str, list[str]], default: None) – Retrieve information about these facets (attributes & values). If None given (default), information about all available facets is returned.
multiversion (bool, default: False) – Select all versions and not just the latest version (default).
**search_facets (str) – The facets to be applied in the data search. If not given the whole dataset will be queried.
- Returns:
Dictionary with a list search facet values for each search facet key.
- Return type:
dict[str, list[str]]
Example
Code
import freva all_facets = freva.facet_search(project='obs*') print(all_facets) spec_facets = freva.facet_search(project='obs*', facet=["time_frequency", "variable"]) print(spec_facets)
Results
{'fs_type': ['posix'], 'cmor_table': ['30min'], 'grid_label': ['gn'], 'realm': ['atmos'], 'institute': ['cpc'], 'time_aggregation': ['mean'], 'future': [], 'variable': ['pr'], 'model': ['cpc'], 'ensemble': ['r1i1p1'], 'project': ['observations'], 'future_id': [], 'experiment': ['cmorph'], 'time_frequency': ['30min'], 'product': ['grid'], 'bbox': []} {'time_frequency': ['30min'], 'variable': ['pr']}Get all models that have a given time step:
Code
import freva model = list(freva.facet_search(project="obs*", time="2016-09-02T22:10")) print(model)
Results
['fs_type', 'cmor_table', 'grid_label', 'realm', 'institute', 'time_aggregation', 'future', 'variable', 'model', 'ensemble', 'project', 'future_id', 'experiment', 'time_frequency', 'product', 'bbox']
Reverse search: retrieving meta data from a known file.
Code
import os, freva file = "../.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020000-201609020030.nc" res = freva.facet_search(file=str(os.path.abspath(file))) print(res)
Results
{'fs_type': ['posix'], 'cmor_table': ['30min'], 'grid_label': ['gn'], 'realm': ['atmos'], 'institute': ['cpc'], 'time_aggregation': ['mean'], 'future': [], 'variable': ['pr'], 'model': ['cpc'], 'ensemble': ['r1i1p1'], 'project': ['observations'], 'future_id': [], 'experiment': ['cmorph'], 'time_frequency': ['30min'], 'product': ['grid'], 'bbox': []}
Running analysis plugins#
Already defined data analysis tools can be started with the freva.run_plugin()
method. Besides the freva.run_plugin() method three more utility methods
(freva.list_plugins(), freva.get_tools_list(),
freva.plugin_doc()) are available to get an overview over
existing plugins and the documentation of each plugins.
- freva.get_tools_list() HelpStr#
Get a list of plugins with their short description.
- Returns:
String representation of all available plugins.
- Return type:
str
Example
Code
import freva import rich rich.print(freva.get_tools_list())
Results
┏━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Tool ┃ Description ┃ ┡━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ Animator │ Animate data on lon/lat grids │ │ DummyPlugin │ A dummy plugin │ │ DummyPluginFolders │ A dummy plugin with outputdir folder │ └────────────────────┴──────────────────────────────────────┘
- freva.list_plugins() list[str]#
Get the plugins that are available on the system.
- Returns:
List of available Freva plugins.
- Return type:
list[str]
Example
Code
import freva import rich rich.print(freva.list_plugins())
Results
['dummyplugin', 'dummypluginfolders', 'animator']
- freva.plugin_doc(tool_name: str | None) HelpStr#
Display the documentation of a given plugin.
- Parameters:
tool_name – The name of the tool that should be documented.
- Returns:
plugin help string.
- Return type:
str
- Raises:
PluginNotFoundError: – if the plugin name does not exist.
Example
Code
import freva import rich rich.print(freva.plugin_doc("animator"))
Results
Animator (v2022.7.15): Create animations (in gif or mp4 format) This tool creates plots of solr facets and an animation. ┏━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Option ┃ Description ┃ ┡━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩ │ input_file │ NetCDF input file(s), you can choose multiple │ │ │ files separated by a , or use a global pattern for │ │ │ multiple files chose this option only if you don't │ │ │ want Freva to find files by seach facets (default: │ │ │ <null>) │ │ variable │ Variable name (only applicable if you didn't │ │ │ choose an input file) (default: <null>) │ │ project │ Project name (only applicable if you didn't choose │ │ │ an input file) (default: <null>) │ │ product │ Product name (only applicable if you didn't choose │ │ │ an input file) (default: <null>) │ │ experiment │ Experiment name (only applicable if you didn't │ │ │ choose an input file) (default: <null>) │ │ institute │ Institute name (only applicable if you didn't │ │ │ choose an input file) (default: <null>) │ │ model │ Model name (only applicable if you didn't choose │ │ │ an input file) (default: <null>) │ │ time_frequency │ Time frequency name (only applicable if you didn't │ │ │ choose an input file) (default: <null>) │ │ ensemble │ Ensemble name (only applicable if you didn't │ │ │ choose an input file) (default: <null>) │ │ start │ Define the first time step to be plotted, leave │ │ │ blank if taken from data (default: <null>) │ │ end │ Define the last time step to be plotted, leave │ │ │ blank if taken from data (default: <null>) │ │ time_mean │ Select a time interval if time │ │ │ averaging/min/max/sum should be applied along the │ │ │ time axis. This can be D for daily, M for monthly │ │ │ 6H for 6 hours etc. Leave blank if the time axis │ │ │ should not be resampled (default) or set to │ │ │ <em>all</em> if you want to collapse the time │ │ │ axis. (default: <null>) │ │ time_method │ If resampling of the time axis is chosen (default: │ │ │ no) set the method: mean, max or min. <b>Note:</b> │ │ │ This has only an effect if the above parameter for │ │ │ <em>time_mean</em> is set. (default: mean) │ │ lonlatbox │ Set the extend of a rectangular lonlatbox │ │ │ (left_lon, right_lon, lower_lat, upper_lat) │ │ │ (default: <null>) │ │ output_unit │ Set the output unit of the variable - leave blank │ │ │ for no conversion. This can be useful if the unit │ │ │ of the input files should be converted, for │ │ │ example for precipitation. Note: Although many │ │ │ conversions are supported, by using the `pint` │ │ │ conversion library. (default: <null>) │ │ vmin │ Set the minimum plotting range (leave blank to │ │ │ calculate from data 1st decile) (default: <null>) │ │ vmax │ Set the maximum plotting range (leave blank to │ │ │ calculate the 9th decile) (default: <null>) │ │ cmap │ Set the colormap, more information on colormaps is │ │ │ available on the <a │ │ │ href="https://matplotlib.org/stable/tutorials/colo │ │ │ rs/colormaps.html" target=_blank>matplotlib │ │ │ website</a>. (default: RdYlBu_r) │ │ linecolor │ Color of the coast lines in the map (default: k) │ │ projection │ Set the global map projection. Note: this should │ │ │ the name of the cartopy projection method (e.g │ │ │ PlatteCarree for Cylindrical Projection). Pleas │ │ │ refer to <a │ │ │ href="https://scitools.org.uk/cartopy/docs/latest/ │ │ │ crs/projections.html"target=_blank>cartopy │ │ │ website</a> for details. (default: PlateCarree) │ │ proj_centre │ Set center longitude of the global map projection. │ │ │ (default: 50) │ │ pic_size │ Set the size of the picture (in pixel) (default: │ │ │ 1360,900) │ │ plot_title │ Set plot title (default: ) │ │ cbar_label │ Overwrite default colorbar label by this value │ │ │ (default: ) │ │ suffix │ Filetype of the animation (default: mp4) │ │ fps │ Set the frames per seceonds of the output │ │ │ animation. (default: 5) │ │ extra_scheduler_options │ Set additional options for the job submission to │ │ │ the workload manager (, separated). Note: │ │ │ batchmode and web only. (default: --qos=test, │ │ │ --array=20) │ └─────────────────────────┴────────────────────────────────────────────────────┘
- freva.run_plugin(tool_name: str, *, save: bool = False, save_config: str | Path | None = None, show_config: bool = False, scheduled_id: int | None = None, unique_output: bool = True, batchmode: bool = False, caption: str = '', tag: str | None = None, **options: str | float | int | bool) PluginStatus#
Apply an available data analysis plugin.
- Parameters:
tool_name – The name of the plugin that is to be applied.
caption – Set a caption for the results.
save – Save the plugin configuration to default destination.
save_config – Save the plugin configuration.
scheduled_id – Run a scheduled job from database
batchmode – Create a Batch job and submit it to the scheduling system.
unique_output – Append a Freva run id to the output/cache folder(s).
tag – Use git commit hash to specify a specific version of this tool.
- Returns:
Return code, and the return value of the plugin.
- Return type:
tuple
Example
Run a plugin in the foreground.
Code
import freva res = freva.run_plugin("animator", variable="pr", project="obs*") output = res.get_result_paths("plot", "*.*") # Check the plot output
Results
/tmp/animator/run.py:91: SyntaxWarning: invalid escape sequence '\d' for i in re.findall("[-\d]+", unit[n]): Setp 1: Collecting all files Step 2: Opening the netcdf-files, collecting metadata Step 3: Converting Units Step 4: Loading the dataset, getting min/max values Step 5: Creating the animation /tmp/animator/run.py:91: SyntaxWarning: invalid escape sequence '\d' for i in re.findall("[-\d]+", unit[n]): Created animation /tmp/eval_conf/work/runner/freva-ces/output/animator/2117/pr_20160902T0000-20160902T2330.mp4 in 40 secondsRun a plugin in the background. You can interact with the plugin using the
.waitmethod of the :py:class:freva.PluginStatusclass.Code
import freva res = freva.run_plugin("animator", variable="pr", project="observations", batchmode=True) res.wait() # Wait until the plugin has finished
Results
Scheduled job with history id: 2118 You can view the job's status with the command squeue Your job's progress will be shown with the command tail -f /tmp/share/slurm/animator/Animator-3561.out ☀️ Waiting for plugin to finish... ok
This specific plugin has created the following output:
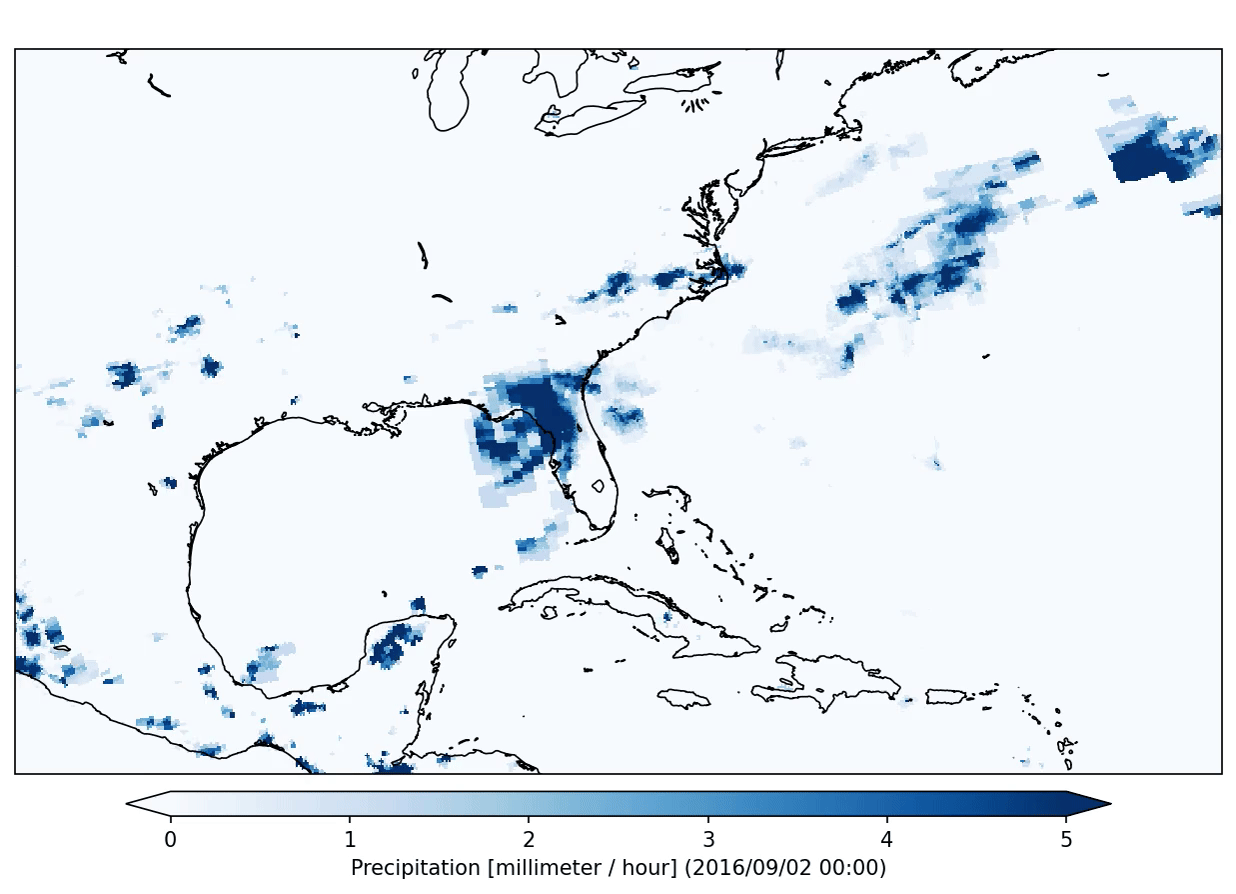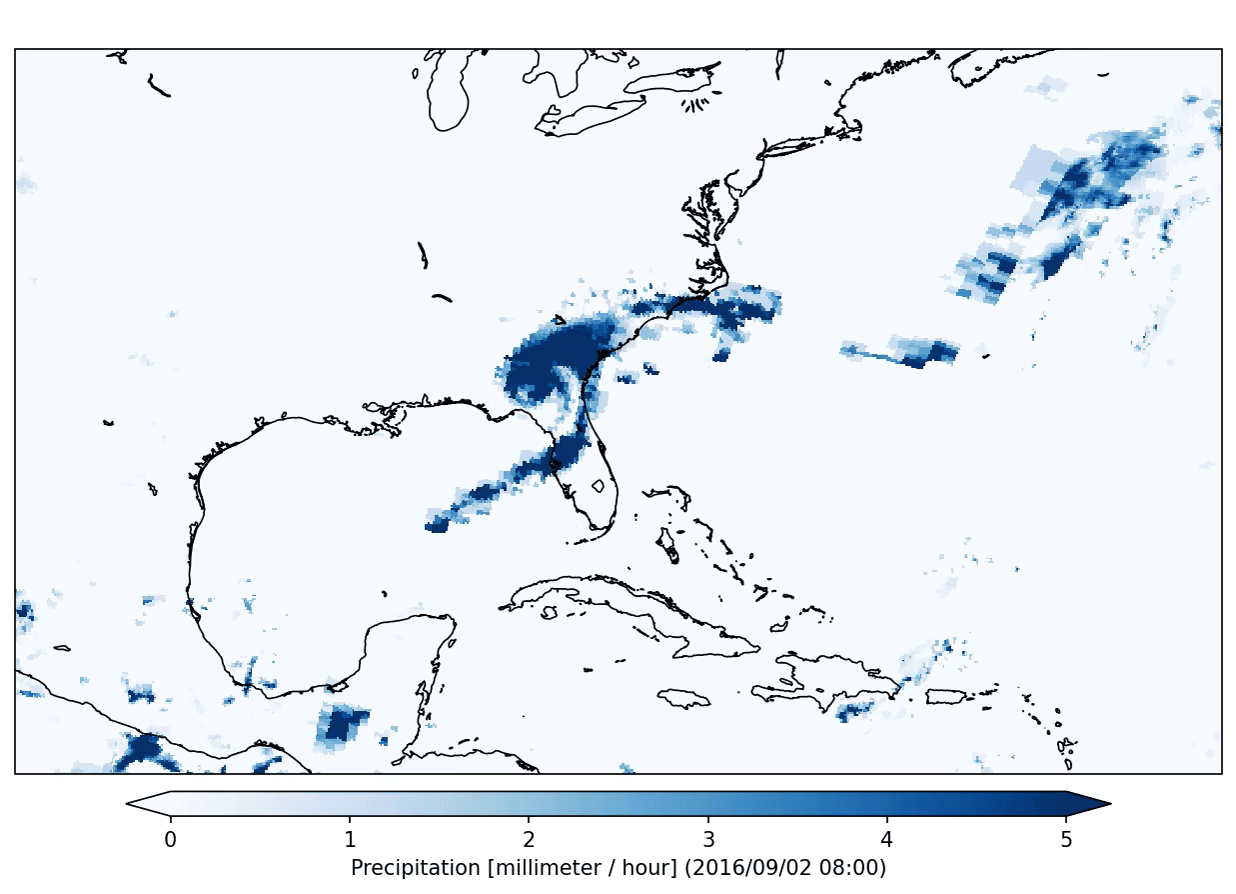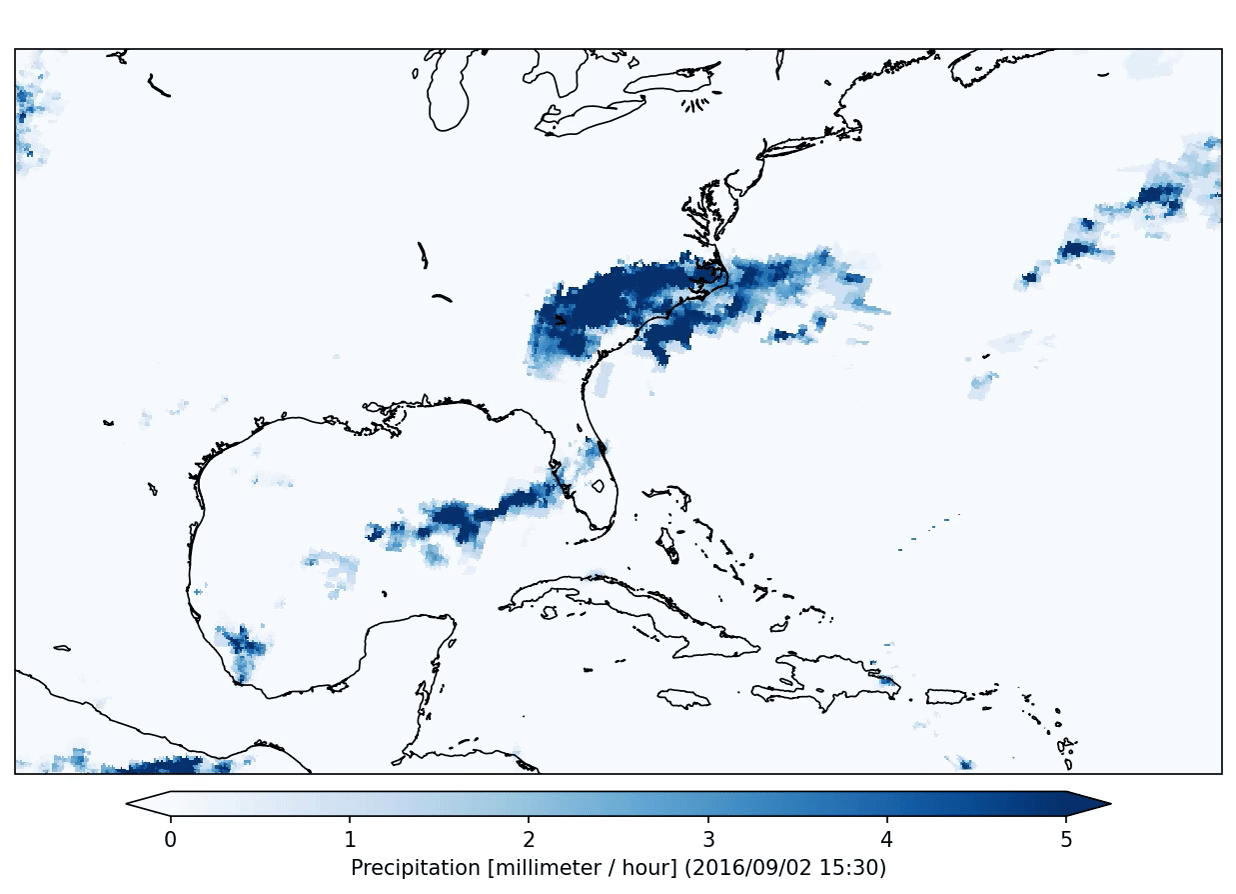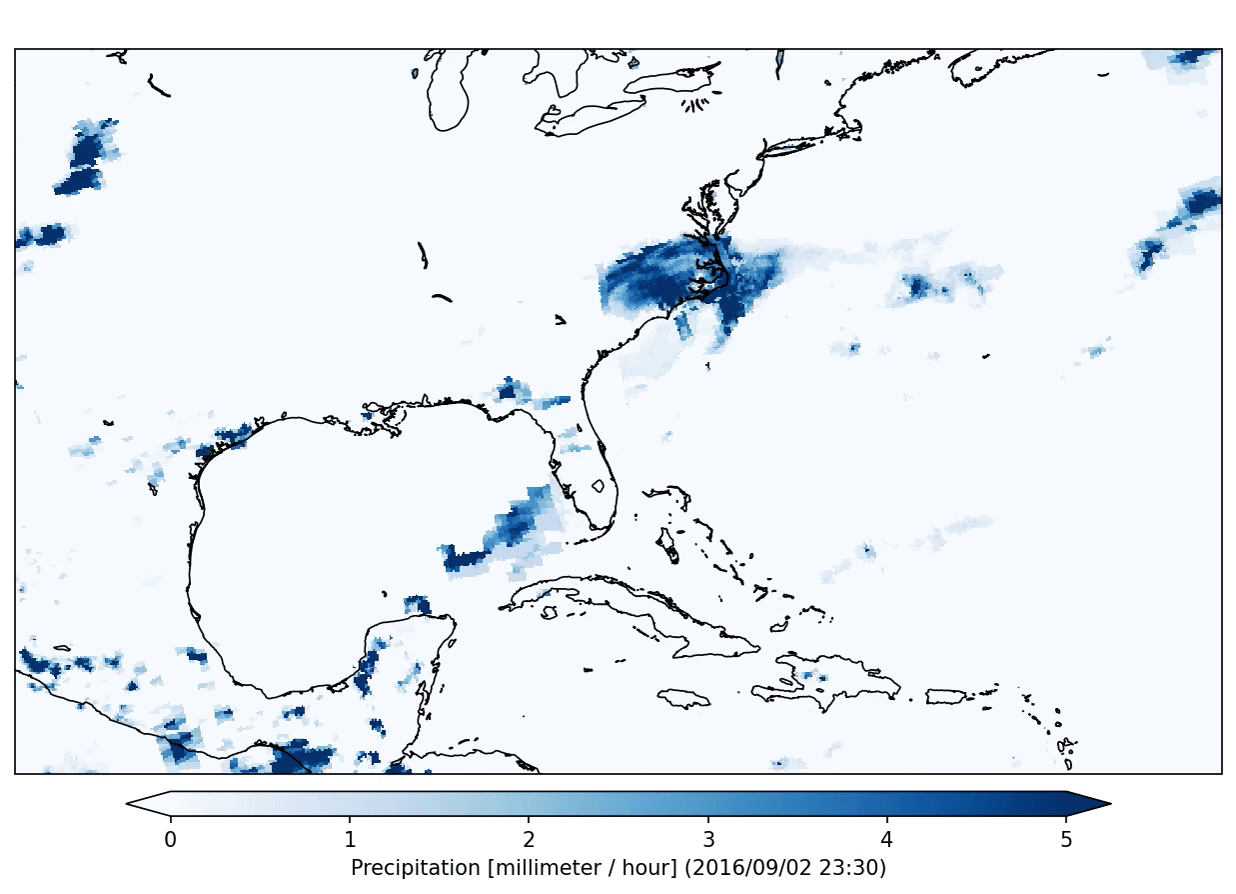
After the application of a data analysis plugin you can check the status of the
plugin and make use of any of the plugin output either by directly using the
return value of the freva.run_plugin() method or create an instance
of the freva.PluginStatus status class with help of a history id.
- class freva.PluginStatus(history_id: int)#
Bases:
objectA class to interact with the status of a plugin application.
With help of this class you can:
Check if a plugin is still running.
Get all results (data or plot files) of a plugin.
Check the configuration of a plugin.
Wait until the plugin is finished.
Example
The output of the
freva.run_pluginmethod is an instance of thePluginStatusclass. That means you can directly use the output of :py:meth:freva.run_pluginto interact with the plugin status:Code
import freva res = freva.run_plugin("dummypluginfolders") print(res.status)
Results
Processing output in /tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2119 finished
You can also create an instance of the class yourself, if you know the
history_idof a specific plugin run. Note that you can query these ids by making use of the :py:meth:freva.historymethod:Code
import freva # Get the last run of the dummypluginfolders plugin hist = freva.history(plugin="dummypluginfolders", limit=1)[:-1] res = freva.PluginStatus(hist["id"]) print(res.status)
Results
[04:47:04] INFO freva - INFO - history of {plugin}, limit=1, since=None, until=None, entry_ids=NoneNote: You won’t be able to kill running jobs associated to history id’s from other users.
- property batch_id: int | None#
Get the id of the batch job, if the plugin was a batchmode job.
- property configuration: Dict[str, Any]#
Get the plugin configuration.
- get_result_paths(dtype: Literal['data', 'plot'] = 'data', glob_pattern: str = '*.nc') List[Path]#
Get all created paths of a certain data type.
This method allows you to query all output files of the plugin run. You can either search for data files or plotted output.
- Parameters:
- Returns:
List[Path]
- Return type:
A list of paths matching the search constrains.
Example
We are going to use a plugin called
dummypluginfolderswhich creates plots and netCDF files. In this example we want to open all netCDF files (dtype = 'data') that match the filename constraint*data.nc.Code
import freva import xarray as xr res = freva.run_plugin("dummypluginfolders", variable="pr") dset = xr.open_mfdataset( res.get_result_paths(dtype="data", glob_pattern="*data.nc") ) print(dset.attrs["variable"])
Results
Processing output in /tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2120 pr
- property history_id: int#
Get the ID of this plugin run in the freva history.
- property job_script: str#
Get the content of the job_script, if it was a batchmode job.
- kill() None#
Kill a running batch job.
This method has only affect on jobs there have been submitted using the
batchmode=Trueflag.
- property plugin: str#
Get the plugin name.
- property status: str#
Get the state of the current plugin run.
- property stdout: str#
Get the stdout of the plugin.
Example
Read the output of the plugin:
Code
import freva res = freva.run_plugin("dummypluginfolders") print(res.stdout)
Results
Processing output in /tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2121 Processing output in /tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2121
- property version: Tuple[int, int, int]#
Get the version of the plugin.
- wait(timeout: float | int = 28800) None#
Wait for a plugin to finish.
This method will block until the plugin is running.
- Parameters:
timeout (int, default: 28800) – Wait
timeoutseconds for the plugin to finish. If the plugin hasn’t been finish raise a ValueError.- Raises:
ValueError – If the plugin took longer than
timeoutseconds to finish.:
Example
This can be useful if a plugin was started using the
batchmode=Trueoption and the execution of the code should wait until the plugin is finished.Code
import freva res = freva.run_plugin("dummypluginfolders", batchmode=True) res.wait(timeout=60) # Give the plugin 60 seconds to finish.
Results
Scheduled job with history id: 2122 You can view the job's status with the command squeue Your job's progress will be shown with the command tail -f /tmp/share/slurm/dummypluginfolders/DummyPluginFolders-3741.out ☀️ Waiting for plugin to finish... ok
Accessing the previous plugin runs#
- freva.history(*args: str, limit: int = 10, plugin: str | None = None, since: str | None = None, until: str | None = None, entry_ids: int | list[int] | None = None, full_text: bool = False, return_results: bool = False, return_command: bool = False, _return_dict: bool = True, user_name: str | None = None) list[Any] | dict[str, Any]#
Get access to the configurations of previously applied freva plugins.
The .history method displays the entries with a one-line compact description. The first number you see is the entry id, which you might use to select single entries.
- Parameters:
limit (int, default: 10) – Limit the number of entries to be displayed.
plugin (str, default: None) – Display only entries from a given plugin name.
since (str, datetime.datetime, default: None) – Retrieve entries older than date, see hint on date format below.
until (str, datetime.datetime, default: None) – Retrieve entries younger than date, see hint on date format below.
entry_ids (list, default: None) – Select entries whose ids are in “ids”.
full_text (bool, default: False) – Show the complete configuration.
return_results (bool, default: False) – Also return the plugin results.
return_command (bool, default: False) – Return the commands instead of history objects.
user_name (str, default: None) – Select entries belonging to another user (e.g., user_name=”<username>”) or all users (all or *).
_return_dict (bool, default: True) – Return a dictionary representation, this is only for internal use.
- Returns:
freva plugin history
- Return type:
list
Example
Get the last three history entries:
Code
import freva hist = freva.history(limit=3) print(type(hist), len(hist)) print(hist[-1].keys()) config = hist[-1]['configuration'] print(config)
Results
<class 'list'> 3 dict_keys(['status_dict', 'id', 'timestamp', 'tool', 'version', 'version_details_id', 'configuration', 'slurm_output', 'host', 'uid_id', 'status', 'flag', 'caption', 'result']) {'variable': 'pr', 'outputdir': '/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704'} [04:47:06] INFO freva - INFO - history, limit=3, since=None, until=None, entry_ids=NoneHint
Date Format Dates are given in the ISO-8601 format and can be “YYYY-MM-DDTHH:mm:ss.n” or any less accurate subset. These are all valid: “2012-02-01T10:08:32.1233431”, “2012-02-01T10:08:32”, “2012-02-01T10:08”, “2012-02-01T10”, “2012-02-01”, “2012-02”, “2012”. Missing values are assumed to be the minimal allowed value. For example: “2012” = “2012-01-01T00:00:00.0”
The UserData class#
- class freva.UserData#
Bases:
objectData class that handles user data requests. With help of this class users can add their own data to the databrowser, (re)-index data in the databrowser or delete data in the databrowser.
- add(product: str, *paths: PathLike, how: str = 'copy', override: bool = False, **defaults: str) None#
Add custom user files to the databrowser.
To be able to add data to the databrowser the file names must follow a strict standard and the files must reside in a specific location. This
addmethod takes care about the correct file naming and location. No pre requirements other than the file has to be a validnetCDForgribfile are assumed. In other words this method places the user data with the correct naming structure to the correct location.- Parameters:
product (str) – Product search key the newly added data can be found.
*paths (os.PathLike) – Filename(s) or Directories that are going to be added to the databrowser. The files will be added into the central user directory and named according the CMOR standard. This ensures that the data can be added into the databrowser. Note: Once the data has been added into the databrowser it can be found via the
user-<username>project.how (str, default: copy) – Method of how the data is added into the central freva user directory. Default is copy, which means your data files will be replicated. To avoid a this redundancy you can set the
howkeyword tosymlinkfor symbolic links orlinkfor creating hard links to create symbolic links ormoveto move the data into the central user directory entirely.override (bool, default: False) – Replace existing files in the user data structure.
experiment (str, default: None) – By default the method tries to deduce the experiment information from the metadata. To overwrite this information the experiment keyword should be set.
institute (str, default: None) – By default the method tries to deduce the institute information from the metadata. To overwrite this information the institute keyword should be set.
model (str, default: None) – By default the method tries to deduce the model information from the metadata. To overwrite this information the model keyword should be set.
variable (str, default: None) – By default the method tries to deduce the variable information from the metadata. To overwrite this information the variable keyword should be set.
time_frequency (str, default: None) – By default the method tries to deduce the time_frequency information from the metadata. To overwrite this information the time_frequency keyword should be set.
ensemble (str, default: None) – By default the method tries to deduce the ensemble information from the metadata. To overwrite this information the ensemble keyword should be set.
- Raises:
ValueError – If metadata is insufficient, or product key is empty.:
Example
Suppose you’ve gotten data from somewhere and want to add this data into the databrowser to make it accessible to others. In this specific example we assume that you have stored your original data in the
/tmp/my_awesome_datafolder. E.g/tmp/my_awesome_data/outfile_0.nc...tmp/my_awesome_data/outfile_9.ncThe routine will try to gather all necessary metadata from the files. You’ll have to provide additional metadata if mandatory keywords are missing. To make the routine work you’ll have to provide theinstitute,modelandexperimentkeywords:Code
from freva import UserData, databrowser user_data = UserData() # You can also provide wild cards to search for data user_data.add("eur-11b", "/tmp/my_awesome_data/outfile_?.nc", institute="clex", model="UM-RA2T", experiment="Bias-correct") # Check the databrowser if the data has been added for file in databrowser(experiment="bias*"): print(file)
Results
Status: crawling ...ok /tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001041800-197001050300.nc /tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001040800-197001041700.nc /tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001032200-197001040700.nc /tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001031200-197001032100.nc /tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001030200-197001031100.nc /tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001021600-197001030100.nc /tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001020600-197001021500.nc /tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001012000-197001020500.nc /tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001011000-197001011900.nc /tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001010000-197001010900.nc
By default the data is copied. By using the
howkeyword you can also link or move the data.
- delete(*paths: PathLike, delete_from_fs: bool = False) None#
Delete data from the databrowser.
The methods deletes user data from the databrowser.
- Parameters:
*paths (os.PathLike) – Filename(s) or Directories that are going to be from the databrowser.
delete_from_fs (bool, default : False) – Do not only delete the files from the databrowser but also from their central location where they have been added to.
- Raises:
ValidationError: – If crawl_dirs do not belong to current user.
Example
Any data in the central user directory that belongs to the user can be deleted from the databrowser and also from the central data location:
Code
from freva import UserData user_data = UserData() user_data.delete(user_data.user_dir)
Results
- index(*crawl_dirs: PathLike, dtype: str = 'fs', continue_on_errors: bool = False, **kwargs: bool) None#
Index and add user output data to the databrowser.
This method can be used to update the databrowser for existing user data.
- Parameters:
crawl_dirs – The data path(s) that needs to be crawled.
dtype – The data type, currently only files on the file system are supported.
continue_on_errors – Continue indexing on error.
- Raises:
ValidationError: – If crawl_dirs do not belong to current user.
Example
If data has been removed from the databrowser it can be re added using the
indexmethod:Code
from freva import UserData user_data = UserData() user_data.index()
Results
Status: crawling ...ok
- property user_dir: Path#
Get the user data directory for the user.
Overriding or using a freva configuration#
If you want to install and maintain an instance of the freva client in your
own python environment you will most likely have to load the freva configuration
to be able to use the freva infrastructure. To do so you can use the
freva.config class. This class allows you to either override or
set the path to the freva configuration file.
- class freva.config(config_file: str | Path | None = None, plugin_path: str | List[str] | None = None)#
Override the default or set the freva system configuration file.
With the help of this class you can not only (temporarily) override the default configuration file and use a configuration from another project, but you can also set a path to a configuration file if no configuration file has been set. Additionally you can set any plugin paths that are not part of the configuration file.
- Parameters:
config_file (str | pathlib.Path, default: None) – Path to the (new) configuration file.
plugin_path (str | List[str], default: None) – New plugins that should be used, use a list of paths if you want export multiple plugins.
Examples
Temporarily override the existing configuration file and use a new one. You can use a context manager to temporally use a different configuration and switch back later.
import freva with freva.config("/work/freva/evaluation_system.conf"): freva.run_plugin("plugin_from_another_project")
If you do not want to switch to another configuration only temporarily, but want to use it permanently, you can use
freva.configwithout a context manager: a context manager:import freva freva.config("/work/freva/evaluation_system.conf") files = sorted(freva.databrowser(project="user-1234", experiment="extremes"))
Import a new user defined plugin, for example if you have created a plugin called
MyPluginthat is located in~/freva/myplugin/plugin.pyyou would set toplugin_path='~/freva/my_plugin,plugin_module'.import freva freva.config(plugin_path="~/freva/my_plugin,plugin_module") freva.run_plugin('MyPlugin", variable1=1, variable2="a")
In the same fashion you can set multiple plugin paths:
- ::
import freva freva.config(plugin_path=[“~/freva/my_plugin1,plugin_module_b”],
“~/ freva/my_plugin2,plugin_module_b”])
- db_reloaded: List[bool] = [False]#
Searching for ESGF data#
Freva also allows to query for data in all the ESGF nodes. You have the following 5 methods:
freva.esgf_browser(): The main method for searching data. The browser method lets you search for data URLs (default) or their opendap/gridftp endpoints.freva.esgf_facets(): This method lists all search categories (facets) and their values.freva.esgf_datasets(): This method lists the name of the datasets (and version) in the ESGF.freva.esgf_download(): You can download the data with the script file that this method creates.freva.esgf_query(): Similarly tofreva.esgf_facets(), this method lists all query elements but groups them by dataset.
Note
The collection of methods
are derived from ESGF’s rest API.
The query facets follow the syntax of the datasets hosted there (CMIP5, CORDEX, CMIP6, etc.)
that might differ from freva.databrowser() and are case
sensitive. They also have some special query keys, e.g.:
distrib: (true, false) search globally or only locally (e.g. at DKRZ, MPI data and replicas)latest: (true, false, unset) search for the latest version, older ones or all.replica: (true, false, unset) search only for replicas, non-replicas, or all.
Below you can find a more detailed documentation.
- freva.esgf_browser(opendap: bool = False, gridftp: bool = False, **search_constraints: dict[str, str]) list[str]#
Find data in the ESGF.
The method queries the ESGF nodes for file URLs (default) or opendap/gridftp endpoints as well.
The
key=valuesyntax follows that offreva.databrowserbut the key names follow the ESGF standards for each dataset. Search of multiple values for the same key can be achieved either as astrconcatenation (e.g.``frequency=”3hr,mon” `` with no space between variables) or as alist(e.g.frequency=["3hr", "mon"]).Parameters:#
- opendap: bool, default: False
List opendap endpoints instead of http ones.
- gridftp: bool, default: False
Show gridftp endpoints instead of the http default ones (or skip them if none found)
- **search_constraints: Union[str, Path, in, list[str]]
Search facets to be applied in the data search.
Returns:#
- list :
Collection of files
Example
Similarly to
freva.databrowser,freva.esgf_browserexpects a list ofkey=valuepairs in no particular order, but unlike the former it is case sensitive.Given that your Freva instance is configured at DKRZ, if we want to search the URLs of all the files stored at the (DKRZ) local node (
distrib=false) holding the latest version (latest=true) of the variable uas (variable=uas) for either 3hr or monthly time frequencies (frequency=["3hr", "mon"]) and for a particular realization within the projectCMIP6:Code
import freva files = freva.esgf_browser( mip_era="CMIP6", activity_id="ScenarioMIP", source_id="CNRM-CM6-1", institution_id="CNRM-CERFACS", experiment_id="ssp585", frequency="3hr,mon", variable="uas", variant_label="r1i1p1f2", distrib=False, latest=True, ) print(f"{len(files) =}") for file in files[:5]: print(file)
Results
len(files) =10 http://esgf3.dkrz.de/thredds/fileServer/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/Amon/uas/gr/v20190219/uas_Amon_CNRM-CM6-1_ssp585_r1i1p1f2_gr_201501-210012.nc http://esgf3.dkrz.de/thredds/fileServer/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_201501010130-202412312230.nc http://esgf3.dkrz.de/thredds/fileServer/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_202501010130-203412312230.nc http://esgf3.dkrz.de/thredds/fileServer/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_203501010130-204412312230.nc http://esgf3.dkrz.de/thredds/fileServer/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_204501010130-205412312230.nc
In order to list the opendap endpoints (
opendap=True):Code
import freva opendap = freva.esgf_browser( mip_era="CMIP6", activity_id="ScenarioMIP", source_id="CNRM-CM6-1", institution_id="CNRM-CERFACS", experiment_id="ssp585", frequency="3hr", variable="uas", variant_label="r1i1p1f2", distrib=False, latest=True, opendap=True, ) print(opendap)
Results
['http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_201501010130-202412312230.nc.html', 'http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_202501010130-203412312230.nc.html', 'http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_203501010130-204412312230.nc.html', 'http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_204501010130-205412312230.nc.html', 'http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_205501010130-206412312230.nc.html', 'http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_206501010130-207412312230.nc.html', 'http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_207501010130-208412312230.nc.html', 'http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_208501010130-209412312230.nc.html', 'http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_209501010130-210012312230.nc.html']
Or the gridftp endpoints instead (
gridftp=True):Results
['gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_201501010130-202412312230.nc', 'gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_202501010130-203412312230.nc', 'gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_203501010130-204412312230.nc', 'gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_204501010130-205412312230.nc', 'gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_205501010130-206412312230.nc', 'gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_206501010130-207412312230.nc', 'gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_207501010130-208412312230.nc', 'gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_208501010130-209412312230.nc', 'gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_209501010130-210012312230.nc']
- freva.esgf_datasets(**search_constraints: dict[str, str]) list[tuple[str, str]]#
List the name of the datasets (and version) in the ESGF.
The method queries the ESGF nodes for dataset information. The
key=valuesyntax follows that offreva.databrowserbut the key names follow the ESGF standards for each dataset.Parameters:#
- **search_constraints: Union[str, Path, in, list[str]]
Search facets to be applied in the data search.
Returns:#
- list[tuple[str, str]]:
list of (dataset_name, version_number) tuples
Example
List the datasets corresponding to a query of files stored at the (DKRZ) local node (
distrib=false) holding the latest version (latest=true) for a particular realization within the projectCMIP6:Code
import freva datasets = freva.esgf_datasets( mip_era="CMIP6", activity_id="ScenarioMIP", source_id="CNRM-CM6-1", institution_id="CNRM-CERFACS", experiment_id="ssp585", frequency="3hr", variable="uas", variant_label="r1i1p1f2", distrib=False, latest=True, ) print(f"{len(datasets) =}") for dataset in datasets[:5]: print(dataset)
Results
len(datasets) =1 ('CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.uas.gr', 20190219)
- freva.esgf_download(download_script: str | Path | None = None, **search_constraints: dict[str, str]) str | Path#
Create a script file to download the queried files at ESGF.
The method creates a bash script wrapper of a wget query from ESGF dataset(s) (only http).
Parameters:#
- download_script: Union[str, Path], default: None
Download wget_script for getting the files instead of displaying anything (only http)
- **search_constraints: Union[str, Path, in, list[str]]
Search facets to be applied in the data search.
Returns:#
- Union[str, Path]:
wget_script to download the files (only http).
Example
Create a wget script to download the queried URLs:
Code
import freva freva.esgf_download(project="CMIP5", experiment="decadal1960", variable="tas", distrib=False, latest=True, download_script="/tmp/script.get") with open('/tmp/script.get', 'r') as f: content = f.readlines() print(" ".join(content[:10]))
Results
#!/bin/bash ############################################################################## # ESG Federation download script # # Template version: 1.2 # Generated by esgf-data.dkrz.de - 2025/05/01 04:47:14 # Search URL: https://esgf-data.dkrz.de/esg-search/wget?project=CMIP5&experiment=decadal1960&variable=tas&distrib=False&latest=True&type=File # ############################################################################### # first be sure it's bash... anything out of bash or sh will break
Note
You will need an OpenID account to download the data, for example here.
There is a ESGF PyClient as well.
- freva.esgf_facets(show_facet: str | list[str] | None = None, **search_constraints: dict[str, str]) dict[str, list[str]]#
Search for data attributes (facets) through ESGF.
The method queries the ESGF nodes for available search facets (keys) like model, experiment etc. The
key=valuesyntax follows that offreva.facet_searchbut the key names follow the ESGF standards for each dataset.Parameters:#
- show_facet: Union[str, list[str]], default: None
List all values for the given facet (might be defined multiple times). The results show the possible values of the selected facet according to the given constraints and the number of datasets (not files) that selecting such value as a constraint will result (faceted search)
- **search_constraints: Union[str, Path, in, list[str]]
Search facets to be applied in the data search.
Returns:#
- dict[str, list[str]]:
Collection of facets
Example
List the values of the attributes
variableandtime_frequencystored at the (DKRZ) local node (distrib=false) holding the latest version (latest=true) for a particular realization within the projectCMIP6:Code
import freva facets = freva.esgf_facets( mip_era="CMIP6", activity_id="ScenarioMIP", source_id="CNRM-CM6-1", institution_id="CNRM-CERFACS", experiment_id="ssp585", distrib=False, latest=True, show_facet=["variable", "frequency"]) print(facets)
Results
{'variable': {'abs550aer': 6, 'agesno': 6, 'areacello': 6, 'baresoilFrac': 5, 'bigthetao': 6, 'bigthetaoga': 6, 'c3PftFrac': 1, 'c4PftFrac': 1, 'ccb': 6, 'cct': 6, 'ch4global': 6, 'ci': 6, 'cl': 6, 'cli': 6, 'clivi': 6, 'clt': 12, 'clw': 6, 'clwvi': 6, 'co2mass': 6, 'cropFrac': 6, 'cropFracC3': 6, 'cropFracC4': 6, 'evspsbl': 6, 'evspsblpot': 6, 'evspsblsoi': 6, 'evspsblveg': 6, 'ficeberg': 6, 'friver': 6, 'gpp': 6, 'grassFrac': 6, 'grassFracC3': 5, 'grassFracC4': 6, 'hcont300': 6, 'hfbasin': 6, 'hfbasinpmadv': 6, 'hfds': 6, 'hfdsn': 6, 'hfls': 7, 'hfss': 7, 'hfx': 6, 'hfy': 6, 'htovgyre': 5, 'htovovrt': 6, 'hur': 6, 'hurs': 11, 'hursmax': 6, 'hursmin': 6, 'hus': 13, 'hus850': 1, 'huss': 13, 'intuadse': 6, 'intuaw': 6, 'intvadse': 6, 'intvaw': 6, 'lai': 12, 'landCoverFrac': 1, 'lwp': 1, 'lwsnl': 6, 'masscello': 6, 'masso': 6, 'mc': 6, 'mfo': 6, 'mlotst': 6, 'mlotstmax': 6, 'mlotstmin': 6, 'mrfso': 6, 'mrlso': 6, 'mrro': 6, 'mrros': 6, 'mrsfl': 12, 'mrsll': 10, 'mrso': 12, 'mrsol': 12, 'mrsos': 6, 'mrtws': 6, 'msftyz': 6, 'n2oglobal': 6, 'npp': 6, 'nwdFracLut': 6, 'o3': 6, 'od550aer': 6, 'od550dust': 1, 'od550so4': 1, 'pbo': 6, 'pr': 8, 'prc': 12, 'prrc': 1, 'prsn': 11, 'prveg': 6, 'prw': 7, 'ps': 7, 'psl': 12, 'ptp': 6, 'ra': 6, 'residualFrac': 6, 'rivo': 6, 'rlds': 13, 'rldscs': 6, 'rls': 1, 'rlus': 12, 'rlut': 11, 'rlutaf': 6, 'rlutcs': 6, 'rlutcsaf': 6, 'rsds': 13, 'rsdscs': 6, 'rsdt': 6, 'rsntds': 6, 'rss': 1, 'rsus': 6, 'rsuscs': 5, 'rsut': 6, 'rsutaf': 6, 'rsutcs': 6, 'rsutcsaf': 6, 'sbl': 11, 'sfcWind': 7, 'sfcWindmax': 12, 'sfdsi': 6, 'siage': 1, 'siareaacrossline': 1, 'siarean': 1, 'siareas': 1, 'sicompstren': 1, 'siconc': 7, 'siconca': 2, 'sidconcdyn': 1, 'sidconcth': 1, 'sidmassdyn': 1, 'sidmassevapsubl': 1, 'sidmassgrowthbot': 1, 'sidmassgrowthwat': 1, 'sidmasslat': 1, 'sidmassmeltbot': 1, 'sidmassmelttop': 1, 'sidmasssi': 1, 'sidmassth': 1, 'sidmasstranx': 1, 'sidmasstrany': 1, 'sidragbot': 1, 'siextentn': 1, 'siextents': 1, 'sifb': 1, 'siflcondbot': 1, 'siflcondtop': 1, 'siflfwbot': 1, 'siflfwdrain': 1, 'sifllatstop': 1, 'sifllwutop': 1, 'siflsensupbot': 1, 'siflswdbot': 1, 'siflswdtop': 1, 'siflswutop': 1, 'sihc': 1, 'simass': 1, 'simassacrossline': 1, 'sipr': 1, 'sisali': 1, 'sisaltmass': 1, 'sisnconc': 1, 'sisnhc': 1, 'sisnmass': 1, 'sisnthick': 7, 'sispeed': 2, 'sistrxdtop': 1, 'sistrxubot': 1, 'sistrydtop': 1, 'sistryubot': 1, 'sitempbot': 1, 'sitempsnic': 1, 'sitemptop': 2, 'sithick': 7, 'sitimefrac': 2, 'siu': 6, 'siv': 7, 'sivol': 6, 'sivoln': 1, 'sivols': 1, 'sltbasin': 1, 'sltnortha': 1, 'sltovgyre': 5, 'sltovovrt': 6, 'snc': 12, 'snd': 6, 'sndmassdyn': 1, 'sndmassmelt': 1, 'sndmasssi': 1, 'sndmasssnf': 1, 'sndmasssubl': 1, 'snm': 6, 'snmassacrossline': 1, 'snw': 6, 'so': 6, 'sob': 6, 'soga': 6, 'sos': 6, 'sosga': 6, 't20d': 6, 'ta': 10, 'ta500': 1, 'ta850': 1, 'tas': 13, 'tasmax': 6, 'tasmin': 7, 'tauu': 6, 'tauuo': 6, 'tauv': 6, 'tauvo': 6, 'thetao': 6, 'thetaoga': 6, 'thetaot': 1, 'thkcello': 6, 'tntrl': 1, 'tob': 6, 'tos': 12, 'tosga': 6, 'tossq': 6, 'toz': 6, 'tpf': 6, 'tran': 6, 'treeFrac': 6, 'ts': 6, 'tsl': 6, 'tslsi': 1, 'tsn': 6, 'ua': 12, 'uas': 14, 'umo': 6, 'uo': 6, 'va': 12, 'vas': 14, 'vegFrac': 6, 'vmo': 6, 'vo': 5, 'volo': 6, 'wap': 6, 'wfo': 6, 'wmo': 6, 'wo': 6, 'wtd': 6, 'zg': 13, 'zg500': 1, 'zos': 6, 'zossq': 6, 'zostoga': 6, 'ztp': 6}, 'frequency': {'3hr': 8, '3hrPt': 4, '6hrPt': 4, 'day': 190, 'fx': 6, 'mon': 1053}}
- freva.esgf_query(query: str | list[str] | None = None, **search_constraints: dict[str, str]) list[dict[str, list[str]]]#
Query fields from ESGF and group them per dataset
The method queries fields (e.g. facets) and groups them by dataset in a list of dictionaries.
Parameters:#
- query:
Display results from <list> queried fields
- **search_constraints:
Search facets to be applied in the data search.
Returns:#
- list[dict[str, list[str]]]:
List of dictionaries with the queried elements for each dataset
Example:#
Show the
key=valuepair of selected elements for a search (e.g.query=["url","master_id","distribution","mip_era","activity_id","source_id","variable","product","version"]):Code
import json, freva queries = freva.esgf_query( mip_era="CMIP6", activity_id="ScenarioMIP", source_id="CNRM-CM6-1", institution_id="CNRM-CERFACS", experiment_id="ssp585", frequency="3hr", variant_label="r1i1p1f2", distrib=False, latest=True, query=["url","master_id","distribution", "mip_era","activity_id","source_id", "variable","product","version"], ) print(json.dumps(queries[:2], indent=3))
Results
[ { "version": "20190219", "activity_id": [ "ScenarioMIP" ], "master_id": "CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.vas.gr", "mip_era": [ "CMIP6" ], "product": [ "model-output" ], "source_id": [ "CNRM-CM6-1" ], "url": [ "http://esgf3.dkrz.de/thredds/catalog/esgcet/1586/CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.vas.gr.v20190219.xml#CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.vas.gr.v20190219|application/xml+thredds|THREDDS", "http://esgf3.dkrz.de/las/getUI.do?catid=649F6BBCDD98B0633CF0042D119CC69D_ns_CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.vas.gr.v20190219|application/las|LAS" ], "variable": [ "vas" ], "score": 1.0 }, { "version": "20190219", "activity_id": [ "ScenarioMIP" ], "master_id": "CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.prrc.gr", "mip_era": [ "CMIP6" ], "product": [ "model-output" ], "source_id": [ "CNRM-CM6-1" ], "url": [ "http://esgf3.dkrz.de/thredds/catalog/esgcet/1586/CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.prrc.gr.v20190219.xml#CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.prrc.gr.v20190219|application/xml+thredds|THREDDS", "http://esgf3.dkrz.de/las/getUI.do?catid=649F6BBCDD98B0633CF0042D119CC69D_ns_CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.prrc.gr.v20190219|application/las|LAS" ], "variable": [ "prrc" ], "score": 1.0 } ]
