The Freva command line interface#
This section introduces the usage of the Freva command line interface - cli. The tutorial assumes that you have already access to Freva either because you’ve setup an instance yourself or one has been setup by the Freva admin team. Hence it is assumed that you know how to access Freva. If this is not the case please contact one of your Freva admins for help.
A general usage overview of the available Freva sub-commands is
available via the --help option:
freva --help
Results
usage: freva [-h] [-V] {databrowser,history,user-data,plugin,esgf} ...
Free EVAluation system framework (freva)
positional arguments:
{databrowser,history,user-data,plugin,esgf}
Available sub-commands:
databrowser Find data in the system.
history Read the plugin application history.
user-data Update users project data.
plugin Apply data analysis plugin.
esgf Search/Download ESGF the data catalogue.
options:
-h, --help show this help message and exit
-V, --version show program's version number and exit
To get help for the individual sub-commands use: freva <sub-command> --help
The most common sub-commands are databrowser and plugin. You can
get more help on the actual commands using the sub-commands --help
option, for example getting help on the databrowser usage:
freva databrowser --help
Results
usage: freva databrowser [-h] [--multiversion] [--batch-size BATCH_SIZE]
[--count] [--facet FACET] [--facet-limit FACET_LIMIT]
[--time-select {flexible,strict,file}] [--debug]
[facets ...]
Find data in the system.
positional arguments:
facets Search facet(s). (default: None)
options:
-h, --help show this help message and exit
--multiversion Select not only the latest version. (default: False)
--batch-size BATCH_SIZE
Number of files to retrieve. (default: 5000)
--count Show the number of files for each search result.
(default: False)
--facet FACET Retrieve values of given facet instead of files.
(default: None)
--facet-limit FACET_LIMIT
Limit the number of output facets. (default:
9223372036854775807)
--time-select {flexible,strict,file}
Operator that specifies how the time period is
selected. (default: flexible)
--debug, -v, -d, --verbose
Use verbose output. (default: False)
Note
Instead of using sub-commands you have also the option to use
commands. For example the command freva-databrowser is equivalent to
freva databrowser, freva-user-data to
freva user-data etc.
Searching for data: the freva-databrowser command#
All files available on in the project are scanned and indexed via a data
search server. This allows you to query the server which
responds almost immediately. To search for data you can either use the
freva-databrowser command or the freva databrowser sub-command.
Let’s inspect the help menu of the databrowser sub-command:
freva-databrowser --help
Results
usage: freva databrowser [-h] [--multiversion] [--batch-size BATCH_SIZE]
[--count] [--facet FACET] [--facet-limit FACET_LIMIT]
[--time-select {flexible,strict,file}] [--debug]
[facets ...]
Find data in the system.
positional arguments:
facets Search facet(s). (default: None)
options:
-h, --help show this help message and exit
--multiversion Select not only the latest version. (default: False)
--batch-size BATCH_SIZE
Number of files to retrieve. (default: 5000)
--count Show the number of files for each search result.
(default: False)
--facet FACET Retrieve values of given facet instead of files.
(default: None)
--facet-limit FACET_LIMIT
Limit the number of output facets. (default:
9223372036854775807)
--time-select {flexible,strict,file}
Operator that specifies how the time period is
selected. (default: flexible)
--debug, -v, -d, --verbose
Use verbose output. (default: False)
The databrowser expects a list of key=value pairs. The order of the
pairs doesn’t really matter. Most important is that you don’t need to
split the search according to the type of data you are searching for.
You can search for files within observations, reanalysis and
model data at the same time. Also important is that all queries are
case insensitive. You can also search for attributes themselves
instead of file paths. For example you can search for the list of
variables available that satisfies a certain constraint (e.g. sampled
6hr, from a certain model, etc).
freva-databrowser project=observations variable=pr model=cp*
Results
/tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001041800-197001050300.nc
/tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001040800-197001041700.nc
/tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001032200-197001040700.nc
/tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001031200-197001032100.nc
/tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001030200-197001031100.nc
/tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001021600-197001030100.nc
/tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001020600-197001021500.nc
/tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001012000-197001020500.nc
/tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001011000-197001011900.nc
/tmp/user_data/user-runner/eur-11b/clex/UM-RA2T/Bias-correct/hr/user_data/hr/r0i0p0/v20250501/tas/tas_hr_UM-RA2T_Bias-correct_r0i0p0_197001010000-197001010900.nc
/home/runner/work/freva/freva/.docker/data/reanalysis/reanalysis/NOAA/NODC/OC5/mon/ocean/Omon/r1i1p1/v20250101/hc700/hc700_mon_NODC_OC5_r1i1p1_201201-201212.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022300-201609022330.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022200-201609022230.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022100-201609022130.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022000-201609022030.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609021900-201609021930.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609021800-201609021830.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609021700-201609021730.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609021600-201609021630.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609021500-201609021530.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609021400-201609021430.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609021300-201609021330.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609021200-201609021230.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609021100-201609021130.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609021000-201609021030.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020900-201609020930.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020800-201609020830.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020700-201609020730.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020600-201609020630.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020500-201609020530.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020400-201609020430.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020300-201609020330.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020200-201609020230.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020100-201609020130.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609020000-201609020030.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/GERICS/NCC-NorESM1-M-GERICS-REMO2015-v1/rcp85/3hr/atmos/3hr/r1i1p1/v20181212/pr/pr_3hr_NCC-NorESM1-M-GERICS-REMO2015-v1_rcp85_r1i1p1_200701020130-200701020430.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/GERICS/NCC-NorESM1-M-GERICS-REMO2015-v1/rcp85/3hr/atmos/3hr/r1i1p1/v20181212/pr/pr_3hr_NCC-NorESM1-M-GERICS-REMO2015-v1_rcp85_r1i1p1_200701010130-200701012230.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/CLMcom/MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1/historical/day/atmos/Aday/r1i1p1/v20140515/tas/tas_day_MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1_historical_r1i1p1_194912101200-194912101200.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/CLMcom/MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1/historical/day/atmos/Aday/r1i1p1/v20140515/tas/tas_day_MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1_historical_r1i1p1_194912091200-194912091200.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/CLMcom/MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1/historical/day/atmos/Aday/r1i1p1/v20140515/tas/tas_day_MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1_historical_r1i1p1_194912081200-194912081200.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/CLMcom/MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1/historical/day/atmos/Aday/r1i1p1/v20140515/tas/tas_day_MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1_historical_r1i1p1_194912071200-194912071200.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/CLMcom/MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1/historical/day/atmos/Aday/r1i1p1/v20140515/tas/tas_day_MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1_historical_r1i1p1_194912061200-194912061200.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/CLMcom/MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1/historical/day/atmos/Aday/r1i1p1/v20140515/tas/tas_day_MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1_historical_r1i1p1_194912051200-194912051200.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/CLMcom/MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1/historical/day/atmos/Aday/r1i1p1/v20140515/tas/tas_day_MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1_historical_r1i1p1_194912041200-194912041200.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/CLMcom/MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1/historical/day/atmos/Aday/r1i1p1/v20140515/tas/tas_day_MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1_historical_r1i1p1_194912031200-194912031200.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/CLMcom/MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1/historical/day/atmos/Aday/r1i1p1/v20140515/tas/tas_day_MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1_historical_r1i1p1_194912021200-194912021200.nc
/home/runner/work/freva/freva/.docker/data/cordex/EUR-11/CLMcom/MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1/historical/day/atmos/Aday/r1i1p1/v20140515/tas/tas_day_MPI-M-MPI-ESM-LR-CLMcom-CCLM4-8-17-v1_historical_r1i1p1_194912011200-194912011200.nc
/home/runner/work/freva/freva/.docker/data/CMIP6/CMIP/MPI-M/MPI-ESM1-2-LR/amip/mon/atmos/Amon/r2i1p1f1/v20190815/ua/ua_mon_MPI-ESM1-2-LR_amip_r2i1p1f1_197901-199812.nc
/home/runner/work/freva/freva/.docker/data/CMIP6/CMIP/CSIRO-ARCCSS/ACCESS-CM2/amip/mon/atmos/Amon/r1i1p1f1/v20191108/ua/ua_mon_ACCESS-CM2_amip_r1i1p1f1_197901-201412.nc
There are many more options for defining a value for a given key:
Attribute syntax |
Meaning |
|---|---|
|
Search for files containing exactly that attribute |
|
Search for files containing a value for attribute that starts with the prefix val |
|
Search for files containing a value for attribute that ends with the suffix lue |
|
Search for files containing a value for attribute that has alu somewhere |
|
Search for files containing a value for attribute that matches the given regular expression (yes! you might use any regular expression to find what you want.) |
OR:
|
Search for files containing either value1 OR value2 for the given attribute (note that’s the same attribute twice!) |
|
Search for files containing value1 for attribute1 AND value2 for attribute2 |
|
Search for files NOT containing value |
|
Search for files containing neither value1 nor value2 |
Note
When using * remember that your shell might give it a different meaning (normally it will try to match files with that name) to turn that off you can use backslash (key=*) or use quotes (key=’*’).
In some cases it might be useful to know how many files are found in the
databrowser for certain search constraints. In such cases you can use the
count flag to count the number of found files instead of getting
the files themselves.
freva-databrowser project=observations --count
Results
49
Sometimes it might be useful to subset the data you’re interested in by time. To do so you can use the time search key to subset time steps and whole time ranges. For example let’s get the for certain time range:
freva-databrowser project=observations time='2016-09-02T22:15 to 2016-10'
Results
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022300-201609022330.nc
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022200-201609022230.nc
The default method for selecting time periods is flexible, which means
all files are selected that cover at least start or end date. The
strict method implies that the entire search time period has to be
covered by the files. Using the strict method in the example above would
only yield on file because the first file contains time steps prior to the
start of the time period:
freva-databrowser project=observations time='2016-09-02T22:15 to 2016-10' --time-select strict
Results
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022300-201609022330.nc
Giving single time steps is also possible:
freva-databrowser project=observations time='2016-09-02T22:10'
Results
/home/runner/work/freva/freva/.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/pr_30min_CPC_cmorph_r1i1p1_201609022200-201609022230.nc
Note
The time format has to follow the
ISO-8601 standard. Time ranges
are indicated by the to keyword such as 2000 to 2100 or
2000-01 to 2100-12 and alike. Single time steps are given without the
to keyword.
In multi versioned datasets only the latest version (i.e. highest version
number) is returned by default. In order to retrieve all the available versions
the --multiversion flag must be activated, e.g.:
freva databrowser project=reanalysis --multiversion
Results
/home/runner/work/freva/freva/.docker/data/reanalysis/reanalysis/NOAA/NODC/OC5/mon/ocean/Omon/r1i1p1/v20250101/hc700/hc700_mon_NODC_OC5_r1i1p1_201201-201212.nc
/home/runner/work/freva/freva/.docker/data/reanalysis/reanalysis/NOAA/NODC/OC5/mon/ocean/Omon/r1i1p1/v20200101/hc700/hc700_mon_NODC_OC5_r1i1p1_201201-201212.nc
Querying a specific version from a multi versioned datasets requires the
--multiversion flag in combination with the version special attribute:
freva databrowser project=reanalysis version=20200101 --multiversion
Results
/home/runner/work/freva/freva/.docker/data/reanalysis/reanalysis/NOAA/NODC/OC5/mon/ocean/Omon/r1i1p1/v20200101/hc700/hc700_mon_NODC_OC5_r1i1p1_201201-201212.nc
You might as well want to know about possible values that an attribute
can take after a certain search is done. For this you use the
--facet flag (facets are the attributes used to search for and sub set
the data). For example to see all facets that are available in the
observations project:
freva-databrowser project=observations --facet all
Results
fs_type: posix
ensemble: r0i0p0,r1i1p1,r1i1p1f1,r2i1p1f1
variable: hc700,pr,tas,ua
cmor_table: 1hr,30min,3hr,aday,amon,omon
project: cmip6,cordex,observations,reanalysis,user-runner
bbox:
institute: clex,clmcom,cpc,csiro-arccss,gerics,mpi-m,noaa
product: cmip,eur-11,eur-11b,grid,reanalysis
future:
model: access-cm2,cpc,mpi-esm1-2-lr,mpi-m-mpi-esm-lr-clmcom-cclm4-8-17-v1,ncc-noresm1-m-gerics-remo2015-v1,nodc,um-ra2t
future_id:
grid_label: gn
time_aggregation: mean
realm: atmos,ocean,user_data
experiment: amip,bias-correct,cmorph,historical,oc5,rcp85
time_frequency: 1day,1hr,30min,3hr,mon
Instead of querying all facets to get information on specific ones:
freva-databrowser --facet time_frequency --facet variable project=observations
Results
time_frequency: 30min
variable: pr
You can also retrieve information on how many facets are found by the databrowser by giving the count flag
freva-databrowser --facet time_frequency --facet variable project=observations --count
Results
time_frequency: 30min (24)
variable: pr (24)
In some cases it might be useful to retrieve meta data from a file path this
can be achieved by using the file= search facet:
freva-databrowser file=../.docker/data/observations/grid/CPC/CPC/cmorph/30min/atmos/30min/r1i1p1/v20210618/pr/ --facet all
Results
fs_type: posix
cmor_table: 30min
grid_label: gn
realm: atmos
institute: cpc
time_aggregation: mean
future:
variable: pr
model: cpc
ensemble: r1i1p1
project: observations
future_id:
experiment: cmorph
time_frequency: 30min
product: grid
bbox:
Running data analysis plugins: the freva-plugin command#
Already defined data analysis tools can be started with the
freva-plugin command or the freva plugin sub-command. Let’s
inspect the help menu of the plugin command:
freva-plugin --help
Results
usage: freva plugin [-h] [--repo-version] [--caption CAPTION] [--save]
[--save-config SAVE_CONFIG] [--show-config] [--batchmode]
[--unique-output {true,false,True,False}] [--debug]
[--list-tools] [--doc] [--json] [--wait]
[plugin_name] [tool-options ...]
Apply data analysis plugin.
positional arguments:
plugin_name Plugin name. (default: None)
tool-options Tool options (default: None)
options:
-h, --help show this help message and exit
--repo-version Show the version number from the repository. (default:
False)
--caption CAPTION Set a caption for the results (default: )
--save Save the plugin configuration to default destination.
(default: False)
--save-config SAVE_CONFIG
Save the plugin configuration. (default: None)
--show-config Show the resulting configuration. (default: False)
--batchmode Create a Batch job and submit it to the scheduling
system. (default: False)
--unique-output {true,false,True,False}, --unique_output {true,false,True,False}
Append a Freva run id to the output/cache folder(s).
(default: true)
--debug, -v, -d, --verbose
Use verbose output. (default: False)
--list-tools, --list, -l
Only list the available tools. (default: False)
--doc, --plugin-doc Display plugin documentation (default: False)
--json, -j Display a json representation of the result, this can
beuseful if you want to build shell based pipelines
and wantparse the output with help of `jq`. (default:
False)
--wait, -w Wait for the plugin to finish, this has only an effect
for batch mode execution. (default: False)
As the help menu suggests you can list all available tools using the
-l option (or --list, --list-tools):
freva-plugin -l
Results
┏━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Tool ┃ Description ┃
┡━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩
│ Animator │ Animate data on lon/lat grids │
│ DummyPlugin │ A dummy plugin │
│ DummyPluginFolders │ A dummy plugin with outputdir folder │
└────────────────────┴──────────────────────────────────────┘
This means currently we have two plugins available (animator and
dummyplugin). The general syntax is
freva-plugin <plugin-name> [options] for example to inspect the
documentation of a certain plugin you can use the --doc option.
Here we concentrate on the Animator plugin. A simple plugin that creates
animations of geospatial data. The basic usage of that command can be
retrieved by:
freva-plugin --doc animator
Results
Animator (v2022.7.15): Create animations (in gif or mp4 format) This tool
creates plots of solr facets and an animation.
┏━━━━━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┓
┃ Option ┃ Description ┃
┡━━━━━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━┩
│ input_file │ NetCDF input file(s), you can choose multiple │
│ │ files separated by a , or use a global pattern for │
│ │ multiple files chose this option only if you don't │
│ │ want Freva to find files by seach facets (default: │
│ │ <null>) │
│ variable │ Variable name (only applicable if you didn't │
│ │ choose an input file) (default: <null>) │
│ project │ Project name (only applicable if you didn't choose │
│ │ an input file) (default: <null>) │
│ product │ Product name (only applicable if you didn't choose │
│ │ an input file) (default: <null>) │
│ experiment │ Experiment name (only applicable if you didn't │
│ │ choose an input file) (default: <null>) │
│ institute │ Institute name (only applicable if you didn't │
│ │ choose an input file) (default: <null>) │
│ model │ Model name (only applicable if you didn't choose │
│ │ an input file) (default: <null>) │
│ time_frequency │ Time frequency name (only applicable if you didn't │
│ │ choose an input file) (default: <null>) │
│ ensemble │ Ensemble name (only applicable if you didn't │
│ │ choose an input file) (default: <null>) │
│ start │ Define the first time step to be plotted, leave │
│ │ blank if taken from data (default: <null>) │
│ end │ Define the last time step to be plotted, leave │
│ │ blank if taken from data (default: <null>) │
│ time_mean │ Select a time interval if time │
│ │ averaging/min/max/sum should be applied along the │
│ │ time axis. This can be D for daily, M for monthly │
│ │ 6H for 6 hours etc. Leave blank if the time axis │
│ │ should not be resampled (default) or set to │
│ │ <em>all</em> if you want to collapse the time │
│ │ axis. (default: <null>) │
│ time_method │ If resampling of the time axis is chosen (default: │
│ │ no) set the method: mean, max or min. <b>Note:</b> │
│ │ This has only an effect if the above parameter for │
│ │ <em>time_mean</em> is set. (default: mean) │
│ lonlatbox │ Set the extend of a rectangular lonlatbox │
│ │ (left_lon, right_lon, lower_lat, upper_lat) │
│ │ (default: <null>) │
│ output_unit │ Set the output unit of the variable - leave blank │
│ │ for no conversion. This can be useful if the unit │
│ │ of the input files should be converted, for │
│ │ example for precipitation. Note: Although many │
│ │ conversions are supported, by using the `pint` │
│ │ conversion library. (default: <null>) │
│ vmin │ Set the minimum plotting range (leave blank to │
│ │ calculate from data 1st decile) (default: <null>) │
│ vmax │ Set the maximum plotting range (leave blank to │
│ │ calculate the 9th decile) (default: <null>) │
│ cmap │ Set the colormap, more information on colormaps is │
│ │ available on the <a │
│ │ href="https://matplotlib.org/stable/tutorials/colo │
│ │ rs/colormaps.html" target=_blank>matplotlib │
│ │ website</a>. (default: RdYlBu_r) │
│ linecolor │ Color of the coast lines in the map (default: k) │
│ projection │ Set the global map projection. Note: this should │
│ │ the name of the cartopy projection method (e.g │
│ │ PlatteCarree for Cylindrical Projection). Pleas │
│ │ refer to <a │
│ │ href="https://scitools.org.uk/cartopy/docs/latest/ │
│ │ crs/projections.html"target=_blank>cartopy │
│ │ website</a> for details. (default: PlateCarree) │
│ proj_centre │ Set center longitude of the global map projection. │
│ │ (default: 50) │
│ pic_size │ Set the size of the picture (in pixel) (default: │
│ │ 1360,900) │
│ plot_title │ Set plot title (default: ) │
│ cbar_label │ Overwrite default colorbar label by this value │
│ │ (default: ) │
│ suffix │ Filetype of the animation (default: mp4) │
│ fps │ Set the frames per seceonds of the output │
│ │ animation. (default: 5) │
│ extra_scheduler_options │ Set additional options for the job submission to │
│ │ the workload manager (, separated). Note: │
│ │ batchmode and web only. (default: --qos=test, │
│ │ --array=20) │
└─────────────────────────┴────────────────────────────────────────────────────┘
The parameters are also given as key=values pairs. But not all of the
above parameters are mandatory. Let’s use one project search key and
animate its content.
freva plugin animator project=observations variable=pr cmap=Blues fps=5 output_unit=mm/h vmin=0 vmax=5 suffix=gif
Results
/tmp/animator/run.py:91: SyntaxWarning: invalid escape sequence '\d'
for i in re.findall("[-\d]+", unit[n]):
Setp 1: Collecting all files
Step 2: Opening the netcdf-files, collecting metadata
Step 3: Converting Units
Step 4: Loading the dataset, getting min/max values
Step 5: Creating the animation
/tmp/animator/run.py:91: SyntaxWarning: invalid escape sequence '\d'
for i in re.findall("[-\d]+", unit[n]):
Created animation /tmp/eval_conf/work/runner/freva-ces/output/animator/2123/pr_20160902T0000-20160902T2330.mp4 in 40 seconds
The plugin will produce the following output:
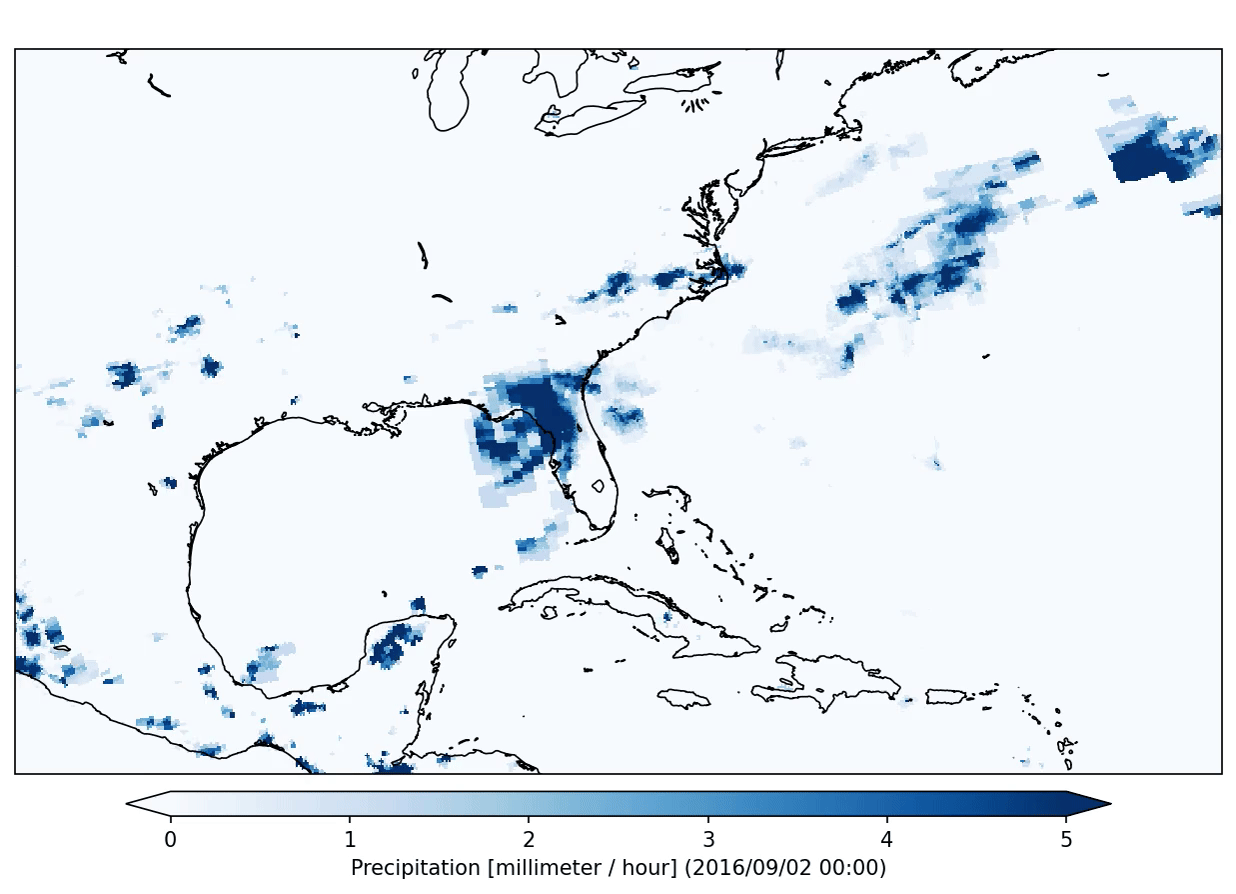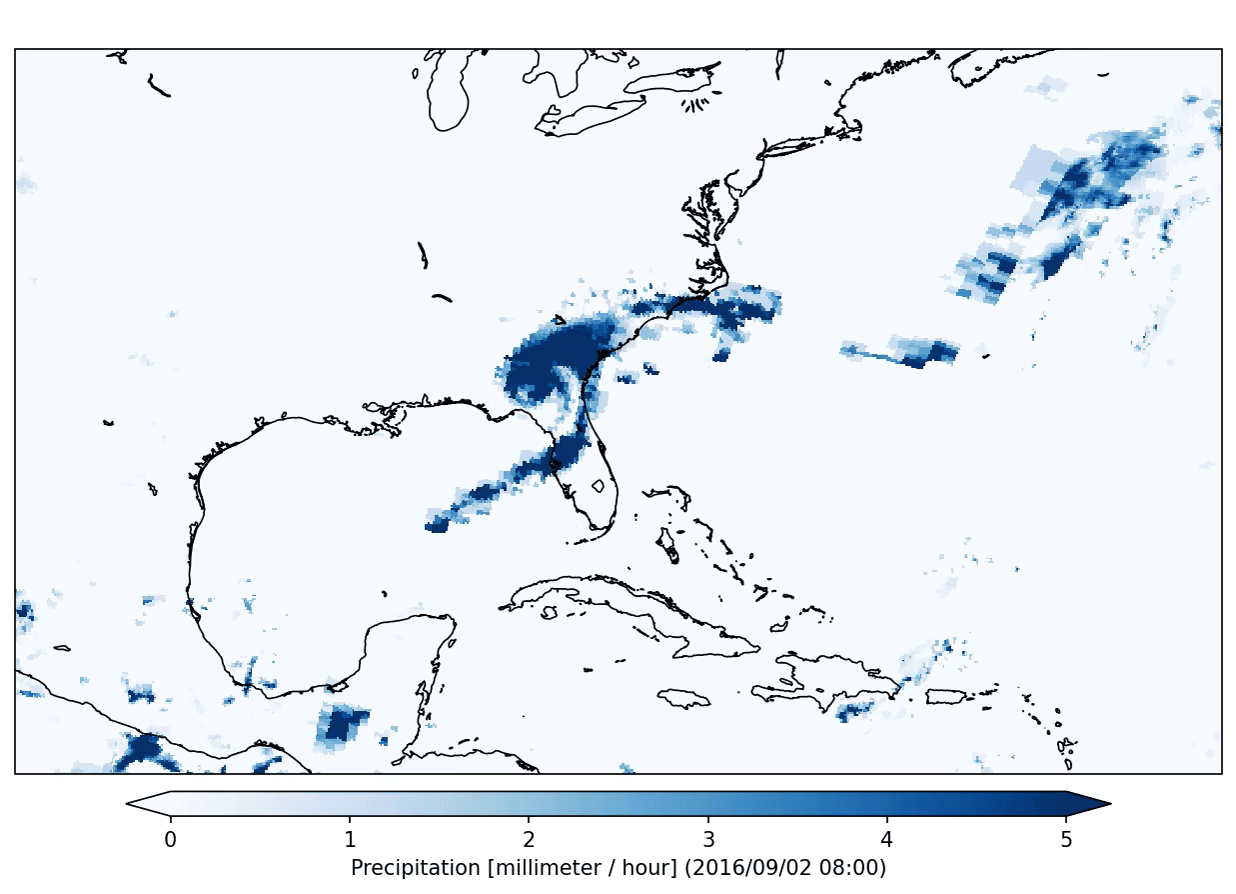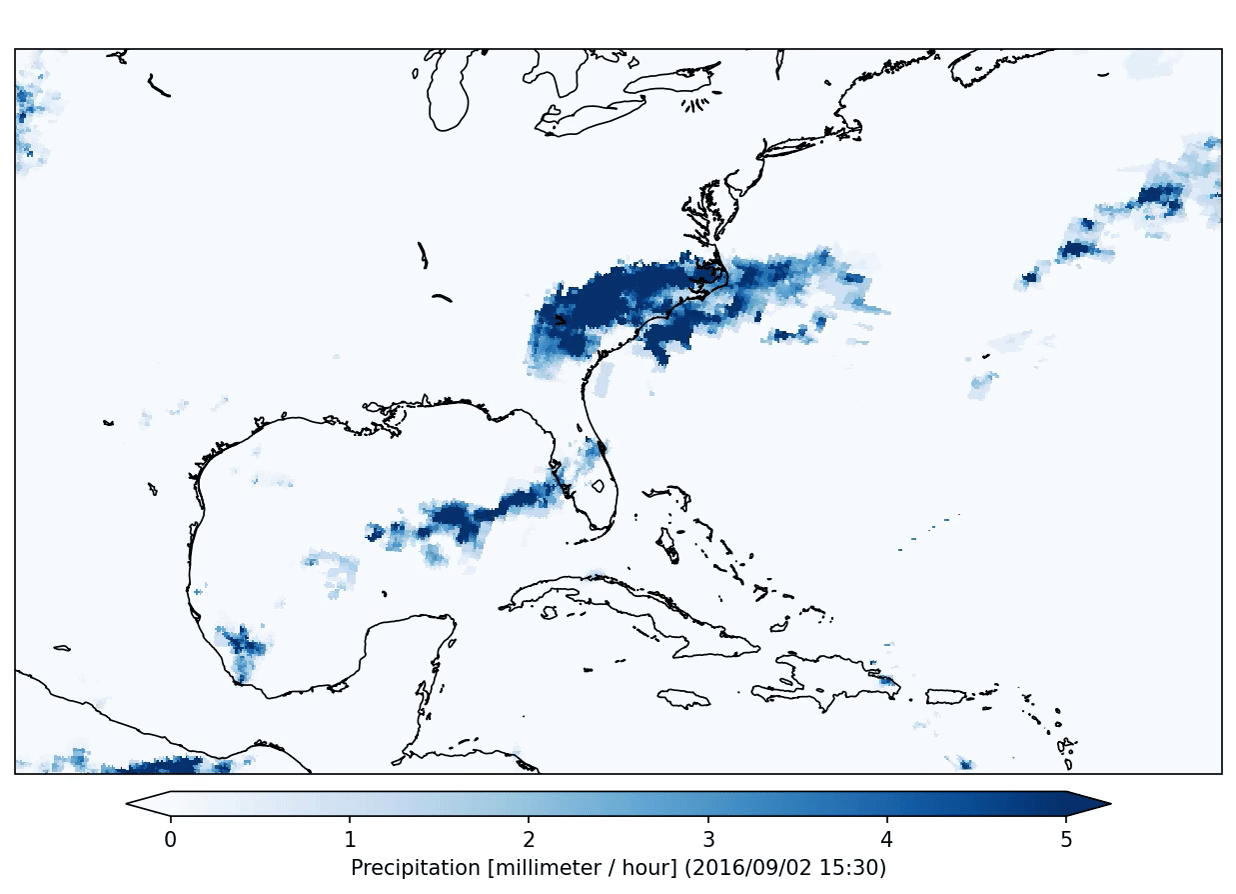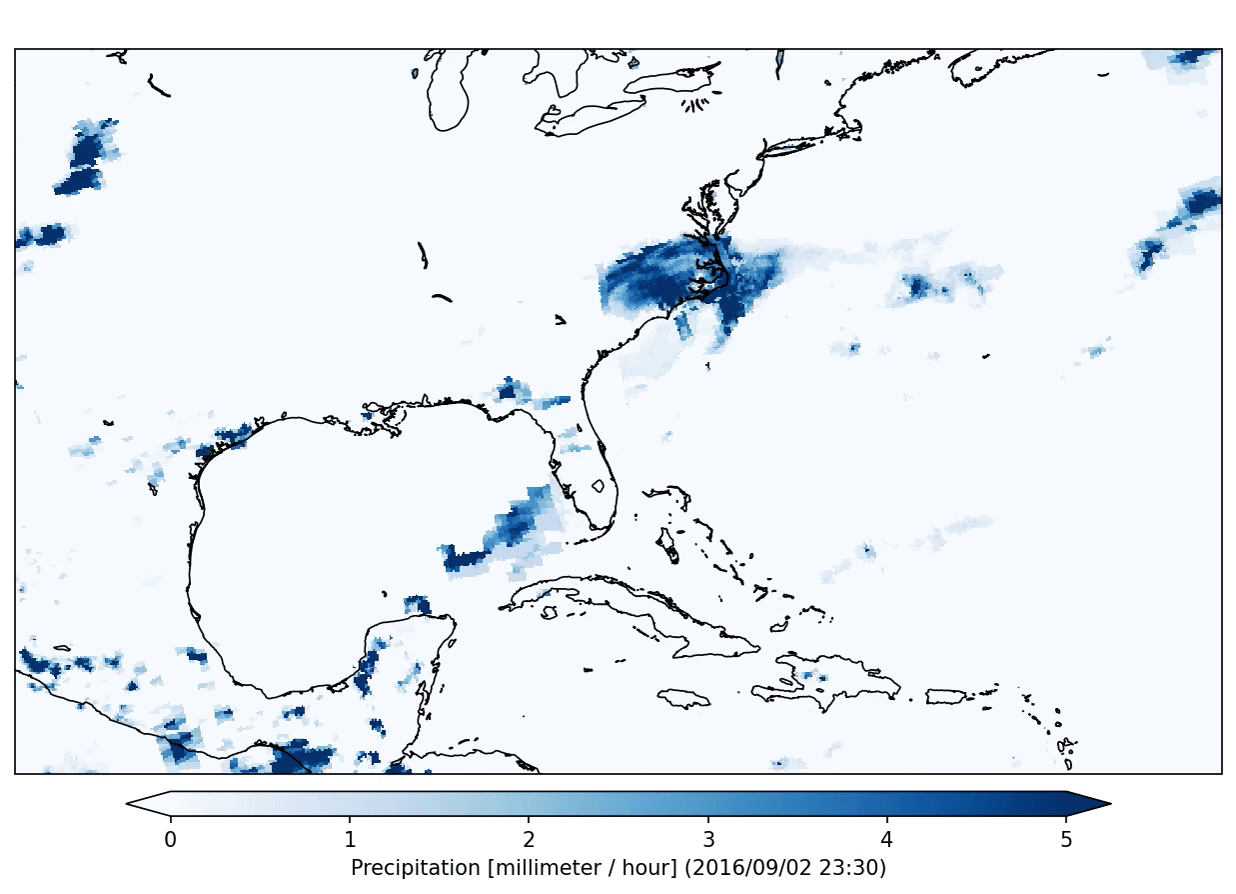
This plugin will run in so called interactive mode. That means that it
will run on the login node and block your shell until the command is
completed. This can be problematic if you have jobs that might take time
to finish. An alternative is setting the -–batchmode flag. This flag
tells the plugin to submit a job to the computing queue. The computing
nodes are the core of any high performance computing system. Let’s
submit the previous plugin job to the computing queue:
freva plugin animator project=observations variable=pr cmap=Blues fps=5 output_unit=mm/h vmin=0 vmax=5 suffix=gif --batchmode
Results
Scheduled job with history id: 2124
You can view the job's status with the command squeue
Your job's progress will be shown with the command
tail -f /tmp/share/slurm/animator/Animator-3977.out
If you want to evaluate the output of the plugin, you can use the --json flag.
This flag converts the result of a plugin application into a json string that
can be processed by a json parser such as jq. Consider the following
example (in bash/zsh) where we use the --json flag to get all the netcdf
output files from a plugin run.
files=$(freva plugin dummypluginfolders --json|jq -r '.result| keys[] | select(endswith(".nc"))')
echo $files
Results
Processing output in /tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817/2125
/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817/2125/data.nc
Note
If you use the --batchmode flag in combination with --json flag the
command line will wait for the batch job to finish.
Inspecting previous analysis jobs: the freva-history command#
Sometimes it can be useful to access the status of past plugin
applications. The freva-history command or freva history
sub-command can do that:
freva-history --help
Results
usage: freva history [-h] [--full-text] [--return-command] [--limit LIMIT]
[--plugin PLUGIN] [--since SINCE] [--until UNTIL]
[--entry-ids ENTRY_IDS [ENTRY_IDS ...]]
[--user-name USER_NAME] [--debug] [--json]
Read the plugin application history.
options:
-h, --help show this help message and exit
--full-text Show the complete configuration. (default: False)
--return-command Show freva commands belonging to the history entries
instead of the entries themselves. (default: False)
--limit LIMIT Limit the number of displayed entries to N. (default:
10)
--plugin PLUGIN Display only entries of selected plugin. (default:
None)
--since SINCE Retrieve entries older than date. (default: None)
--until UNTIL Retrieve entries newer than date. (default: None)
--entry-ids ENTRY_IDS [ENTRY_IDS ...]
Select entry id(s) (e.g. --entry-ids 1 --entry-ids 2
). (default: None)
--user-name USER_NAME
Show the history of other user (e.g. --user-name
<username>) or all users (--user-name all or --user-
name '*'). (default: None)
--debug, -v, -d, --verbose
Use verbose output. (default: False)
--json, -j Display a json representation of the output, this can
beuseful if you want to build shell based pipelines
and wantparse the output with help of `jq`. (default:
False)
Let’s get the last entry (default is 10 entries) of the dummyplugin plugin history
freva-history --limit 1 --plugin dummyplugin
Results
2115) dummyplugin [2025-05-01 04:44:33] finished {"number": null, "the_number": 5, "something": "test", "other": 1.4...
Dates are given using the ISO-8601 format.
The entries are sorted by their id. For example you can query the
full configuration by giving the id:
freva-history --entry-ids 136 --full-text
Results
2125) dummypluginfolders(0, 1, 2) dbc985023b8e54e92584d49889b25ee88435b1c7 [2025-05-01 04:48:17] finished
Configuration:
outputdir=/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817
variable=tas
To re-run a command of a past configuration you
can use the --return-command option to get the command that was used:
freva-history --entry-ids 136 --return-command
Results
freva-plugin dummypluginfolders variable=tas outputdir=/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817
As with the plugin subcommand, you can use the --json flag
to make the output of the history command machine-readable,
and to parse its output. For example, we can query the output files
of the last 3 plugin applications:
freva-history --limit 3 --json|jq -r '.[].result | keys[]'
Results
/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817/2125/data.nc
/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817/2125/plot.png
/tmp/eval_conf/work/runner/freva-ces/output/animator/2123/pr_20160902T0000-20160902T2330.mp4
[04:48:21] INFO freva - INFO - history, limit=3, since=None, until=None,
entry_ids=None
By default, Freva only shows your user’s history. However,
with the --user-name flag, the query can be extended to other users (e.g.
--user-name <username>) or all users (--user-name all or --user-name "*").
For example, while the following Freva command shows the jobs run by you:
freva history --json
Results
Processing output in /tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044821/2126
[
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2126,
"timestamp": "2025-05-01T04:48:21",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "tas",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044821"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-ea776d0d-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044821/2126/data.nc": {
"timestamp": 1746074901,
"size": 255,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044821/2126/plot.png": {
"timestamp": 1746074901,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044821_IXildtdZ.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2125,
"timestamp": "2025-05-01T04:48:17",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "tas",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-302256af-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817/2125/data.nc": {
"timestamp": 1746074897,
"size": 255,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817/2125/plot.png": {
"timestamp": 1746074897,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044817_VpHoNreg.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2124,
"timestamp": "2025-05-01T04:48:16",
"tool": "animator",
"version": "(2022, 7, 15)",
"version_details_id": 124,
"configuration": {
"input_file": null,
"variable": "pr",
"project": "observations",
"product": null,
"experiment": null,
"institute": null,
"model": null,
"time_frequency": null,
"ensemble": null,
"start": null,
"end": null,
"time_mean": null,
"time_method": "mean",
"lonlatbox": null,
"output_unit": "mm/h",
"vmin": 0.0,
"vmax": 5.0,
"cmap": "Blues",
"linecolor": "k",
"projection": "PlateCarree",
"proj_centre": 50,
"pic_size": "1360,900",
"plot_title": "",
"cbar_label": "",
"suffix": "gif",
"fps": 5
},
"slurm_output": "/tmp/share/slurm/animator/Animator-3977.out",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 3,
"flag": 0,
"caption": "",
"result": {}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2123,
"timestamp": "2025-05-01T04:47:33",
"tool": "animator",
"version": "(2022, 7, 15)",
"version_details_id": 124,
"configuration": {
"input_file": null,
"variable": "pr",
"project": "observations",
"product": null,
"experiment": null,
"institute": null,
"model": null,
"time_frequency": null,
"ensemble": null,
"start": null,
"end": null,
"time_mean": null,
"time_method": "mean",
"lonlatbox": null,
"output_unit": null,
"vmin": null,
"vmax": null,
"cmap": "RdYlBu_r",
"linecolor": "k",
"projection": "PlateCarree",
"proj_centre": 50,
"pic_size": "1360,900",
"plot_title": "",
"cbar_label": "",
"suffix": "mp4",
"fps": 5
},
"slurm_output": "/tmp/share/slurm/animator/Animator-c31757bd-3872.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/animator/2123/pr_20160902T0000-20160902T2330.mp4": {
"timestamp": 1746074895,
"size": 1492223,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/animator_20250501_044815_tnERAlIy.mp4"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2122,
"timestamp": "2025-05-01T04:47:05",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "tas",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044705"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-3741.out",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044705/2122/data.nc": {
"timestamp": 1746074826,
"size": 255,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044705/2122/plot.png": {
"timestamp": 1746074826,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044706_njrTPRgL.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2121,
"timestamp": "2025-05-01T04:47:04",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "tas",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-5c64c5c4-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2121/data.nc": {
"timestamp": 1746074824,
"size": 255,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2121/plot.png": {
"timestamp": 1746074824,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044704_jmNiIenA.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2120,
"timestamp": "2025-05-01T04:47:04",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "pr",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-19c339db-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2120/data.nc": {
"timestamp": 1746074824,
"size": 254,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2120/plot.png": {
"timestamp": 1746074824,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044704_yysYvFql.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2119,
"timestamp": "2025-05-01T04:47:04",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "tas",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-11eeaecd-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2119/data.nc": {
"timestamp": 1746074824,
"size": 255,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2119/plot.png": {
"timestamp": 1746074824,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044704_YPgEXRvu.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2118,
"timestamp": "2025-05-01T04:46:20",
"tool": "animator",
"version": "(2022, 7, 15)",
"version_details_id": 124,
"configuration": {
"input_file": null,
"variable": "pr",
"project": "observations",
"product": null,
"experiment": null,
"institute": null,
"model": null,
"time_frequency": null,
"ensemble": null,
"start": null,
"end": null,
"time_mean": null,
"time_method": "mean",
"lonlatbox": null,
"output_unit": null,
"vmin": null,
"vmax": null,
"cmap": "RdYlBu_r",
"linecolor": "k",
"projection": "PlateCarree",
"proj_centre": 50,
"pic_size": "1360,900",
"plot_title": "",
"cbar_label": "",
"suffix": "mp4",
"fps": 5
},
"slurm_output": "/tmp/share/slurm/animator/Animator-3561.out",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/animator/2118/pr_20160902T0000-20160902T2330.mp4": {
"timestamp": 1746074823,
"size": 1492223,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/animator_20250501_044703_FHPJMkbk.mp4"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2117,
"timestamp": "2025-05-01T04:45:37",
"tool": "animator",
"version": "(2022, 7, 15)",
"version_details_id": 124,
"configuration": {
"input_file": null,
"variable": "pr",
"project": "obs*",
"product": null,
"experiment": null,
"institute": null,
"model": null,
"time_frequency": null,
"ensemble": null,
"start": null,
"end": null,
"time_mean": null,
"time_method": "mean",
"lonlatbox": null,
"output_unit": null,
"vmin": null,
"vmax": null,
"cmap": "RdYlBu_r",
"linecolor": "k",
"projection": "PlateCarree",
"proj_centre": 50,
"pic_size": "1360,900",
"plot_title": "",
"cbar_label": "",
"suffix": "mp4",
"fps": 5
},
"slurm_output": "/tmp/share/slurm/animator/Animator-b4ba315e-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/animator/2117/pr_20160902T0000-20160902T2330.mp4": {
"timestamp": 1746074779,
"size": 1492223,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/animator_20250501_044619_NXHHlsRk.mp4"
}
}
}
]
The following one will show you the history of e.g. root user:
freva history --json --user-name=root
Results
[
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2127,
"timestamp": "2025-05-01T04:48:22",
"tool": "new_dummytool",
"version": "",
"version_details_id": 1,
"configuration": {
"some": "config",
"dict": "values"
},
"slurm_output": "/path/to/slurm-44742.out",
"host": "10.1.0.88",
"uid_id": "root",
"status": 0,
"flag": 0,
"caption": "",
"result": {}
}
]
And the following one, of all users (that is, including you):
freva history --json --user-name all
Results
[
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2127,
"timestamp": "2025-05-01T04:48:22",
"tool": "new_dummytool",
"version": "",
"version_details_id": 1,
"configuration": {
"some": "config",
"dict": "values"
},
"slurm_output": "/path/to/slurm-44742.out",
"host": "10.1.0.88",
"uid_id": "root",
"status": 0,
"flag": 0,
"caption": "",
"result": {}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2126,
"timestamp": "2025-05-01T04:48:21",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "tas",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044821"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-ea776d0d-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044821/2126/data.nc": {
"timestamp": 1746074901,
"size": 255,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044821/2126/plot.png": {
"timestamp": 1746074901,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044821_IXildtdZ.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2125,
"timestamp": "2025-05-01T04:48:17",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "tas",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-302256af-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817/2125/data.nc": {
"timestamp": 1746074897,
"size": 255,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044817/2125/plot.png": {
"timestamp": 1746074897,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044817_VpHoNreg.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2124,
"timestamp": "2025-05-01T04:48:16",
"tool": "animator",
"version": "(2022, 7, 15)",
"version_details_id": 124,
"configuration": {
"input_file": null,
"variable": "pr",
"project": "observations",
"product": null,
"experiment": null,
"institute": null,
"model": null,
"time_frequency": null,
"ensemble": null,
"start": null,
"end": null,
"time_mean": null,
"time_method": "mean",
"lonlatbox": null,
"output_unit": "mm/h",
"vmin": 0.0,
"vmax": 5.0,
"cmap": "Blues",
"linecolor": "k",
"projection": "PlateCarree",
"proj_centre": 50,
"pic_size": "1360,900",
"plot_title": "",
"cbar_label": "",
"suffix": "gif",
"fps": 5
},
"slurm_output": "/tmp/share/slurm/animator/Animator-3977.out",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 3,
"flag": 0,
"caption": "",
"result": {}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2123,
"timestamp": "2025-05-01T04:47:33",
"tool": "animator",
"version": "(2022, 7, 15)",
"version_details_id": 124,
"configuration": {
"input_file": null,
"variable": "pr",
"project": "observations",
"product": null,
"experiment": null,
"institute": null,
"model": null,
"time_frequency": null,
"ensemble": null,
"start": null,
"end": null,
"time_mean": null,
"time_method": "mean",
"lonlatbox": null,
"output_unit": null,
"vmin": null,
"vmax": null,
"cmap": "RdYlBu_r",
"linecolor": "k",
"projection": "PlateCarree",
"proj_centre": 50,
"pic_size": "1360,900",
"plot_title": "",
"cbar_label": "",
"suffix": "mp4",
"fps": 5
},
"slurm_output": "/tmp/share/slurm/animator/Animator-c31757bd-3872.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/animator/2123/pr_20160902T0000-20160902T2330.mp4": {
"timestamp": 1746074895,
"size": 1492223,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/animator_20250501_044815_tnERAlIy.mp4"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2122,
"timestamp": "2025-05-01T04:47:05",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "tas",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044705"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-3741.out",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044705/2122/data.nc": {
"timestamp": 1746074826,
"size": 255,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044705/2122/plot.png": {
"timestamp": 1746074826,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044706_njrTPRgL.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2121,
"timestamp": "2025-05-01T04:47:04",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "tas",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-5c64c5c4-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2121/data.nc": {
"timestamp": 1746074824,
"size": 255,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2121/plot.png": {
"timestamp": 1746074824,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044704_jmNiIenA.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2120,
"timestamp": "2025-05-01T04:47:04",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "pr",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-19c339db-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2120/data.nc": {
"timestamp": 1746074824,
"size": 254,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2120/plot.png": {
"timestamp": 1746074824,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044704_yysYvFql.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2119,
"timestamp": "2025-05-01T04:47:04",
"tool": "dummypluginfolders",
"version": "(0, 1, 2)",
"version_details_id": 125,
"configuration": {
"variable": "tas",
"outputdir": "/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704"
},
"slurm_output": "/tmp/share/slurm/dummypluginfolders/DummyPluginFolders-11eeaecd-3336.local",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2119/data.nc": {
"timestamp": 1746074824,
"size": 255,
"type": "data"
},
"/tmp/eval_conf/work/runner/freva-ces/output/dummypluginfolders/20250501_044704/2119/plot.png": {
"timestamp": 1746074824,
"size": 636,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/dummypluginfolders_20250501_044704_YPgEXRvu.png"
}
}
},
{
"status_dict": {
"2": "broken",
"0": "finished",
"1": "finished_no_output",
"5": "not_scheduled",
"3": "running",
"4": "scheduled"
},
"id": 2118,
"timestamp": "2025-05-01T04:46:20",
"tool": "animator",
"version": "(2022, 7, 15)",
"version_details_id": 124,
"configuration": {
"input_file": null,
"variable": "pr",
"project": "observations",
"product": null,
"experiment": null,
"institute": null,
"model": null,
"time_frequency": null,
"ensemble": null,
"start": null,
"end": null,
"time_mean": null,
"time_method": "mean",
"lonlatbox": null,
"output_unit": null,
"vmin": null,
"vmax": null,
"cmap": "RdYlBu_r",
"linecolor": "k",
"projection": "PlateCarree",
"proj_centre": 50,
"pic_size": "1360,900",
"plot_title": "",
"cbar_label": "",
"suffix": "mp4",
"fps": 5
},
"slurm_output": "/tmp/share/slurm/animator/Animator-3561.out",
"host": "fv-az1755-108",
"uid_id": "runner",
"status": 0,
"flag": 0,
"caption": "",
"result": {
"/tmp/eval_conf/work/runner/freva-ces/output/animator/2118/pr_20160902T0000-20160902T2330.mp4": {
"timestamp": 1746074823,
"size": 1492223,
"type": "plot",
"todo": "copy",
"preview_path": "/tmp/share/preview/20250501/animator_20250501_044703_FHPJMkbk.mp4"
}
}
}
]
Managing your own datasets: the freva-user-data command#
Freva offers the possibility to share custom datasets with other users
by making it searchable via freva-databrowser. With help of the
freva-user-data command users can add their own data to the
central data location, (re)-index or delete data in the databrowser.
Note
Any data that has been added by users will be assigned a special project
name: project=user-$USER.
Let’s inspect the help menu of the freva-user-data or freva user-data
command:
freva-user-data --help
Results
usage: freva user-data [-h] {index,add,delete} ...
Update users project data.
positional arguments:
{index,add,delete} Available sub-commands:
index Index existing user project data to the databrowser.
add Add new user project data to the databrowser.
delete Delete existing user project data from the databrowser.
options:
-h, --help show this help message and exit
Add new data to the databrowser#
To be able to add data to the databrowser the file names must follow a strict
standard and the files must reside in a specific location. The
add sub command takes care about the correct file naming and location.
No pre requirements other than the file has to be a valid netCDF or
grib file are assumed. In other words this sub command places the user data
with the correct naming structure to the correct location.
freva-user-data add --help
Results
usage: freva user-data add [-h] [--how {copy,move,symlink,link}] [--override]
[--experiment EXPERIMENT] [--institute INSTITUTE]
[--model MODEL] [--variable VARIABLE]
[--time-frequency TIME_FREQUENCY]
[--ensemble ENSEMBLE] [--cmor-table CMOR_TABLE]
[--realm REALM] [--debug]
product paths [paths ...]
Add new user project data to the databrowser.
positional arguments:
product Product search key the newly added data can be found.
paths Filename(s) or Directories that are going to be added
to thedatabrowser.
options:
-h, --help show this help message and exit
--how {copy,move,symlink,link}
Method of how the data is added into the central freva
user directory. (default: copy)
--override, --overwrite
Replace existing files in the user data structure.
(default: False)
--experiment EXPERIMENT
Set the <experiment> information if they can't be
found in the meta data. (default: None)
--institute INSTITUTE
Set the <institute> information if they can't be found
in the meta data. (default: None)
--model MODEL Set the <model> information if they can't be found in
the meta data. (default: None)
--variable VARIABLE Set the <variable> information if they can't be found
in the meta data. (default: None)
--time-frequency TIME_FREQUENCY, --time_frequency TIME_FREQUENCY
Set the <time_frequency> information if they can't be
found in the meta data. (default: None)
--ensemble ENSEMBLE Set the <ensemble> information if they can't be found
in the meta data. (default: None)
--cmor-table CMOR_TABLE, --cmor_table CMOR_TABLE
Set the <cmor-table> information if they can't be
found in the meta data. (default: None)
--realm REALM Set the <realm> information if they can't be found in
the metadata. (default: None)
--debug, -v, -d, --verbose
Use verbose output. (default: False)
Suppose you’ve gotten data from somewhere and want to add this data into the
databrowser to make it accessible to others. In this specific
example we assume that you have stored your original data in the
/tmp/my_awesome_data folder.
E.g /tmp/my_awesome_data/outfile_0.nc...tmp/my_awesome_data/outfile_9.nc
The routine will try to gather all necessary metadata from the files. You’ll
have to provide additional metadata if mandatory keywords are missing.
To make the routine work in this example we have to provide the institute,
model and experiment keywords:
freva-user-data add eur-11b /tmp/my_awesome_data/outfile_?.nc \
--institute clex --model UM-RA2T --experiment Bias-correct
freva-databrowser experiment=bias-correct
Results
Status: crawling ...ok
Note
Freva allows also plugins to directly index output datasets via
add_output_to_databrowser method (linkmydata is the deprecated
method of former Freva versions). For more information please
take a look at Plugin API Reference.
Remove your data from the databrowser#
The delete sub command removes entries from the databrowser and if necessary
existing files from the central user data location.
freva-user-data delete --help
Results
usage: freva user-data delete [-h] [--delete-from-fs] [--debug]
paths [paths ...]
Delete existing user project data from the databrowser.
positional arguments:
paths The user directory(s) that needs to be crawled.
options:
-h, --help show this help message and exit
--delete-from-fs, --delete_from_fs, --delete
Do not only delete the files from the databrowser but
also from file system. (default: False)
--debug, -v, -d, --verbose
Use verbose output. (default: False)
Any data in the central user directory that belongs to the user can be deleted from the databrowser and also from the central data location:
freva-user-data delete /tmp/user_data/user-<user_name>
freva-databrowser experiment=bias-correct
Results
(Re)-Index existing data to the databrowser#
The index subcommand can be used to update the databrowser for existing
user data. For example, if data has been removed from the databrowser it can
be re-added:
freva-user-data index --help
Results
usage: freva user-data index [-h] [--data-type {fs}] [--continue-on-errors]
[--debug]
[crawl_dir ...]
Index existing user project data to the databrowser.
positional arguments:
crawl_dir The user directory(s) that needs to be crawled.
(default: None)
options:
-h, --help show this help message and exit
--data-type {fs}, --dtype {fs}
The data type of the data. (default: fs)
--continue-on-errors, --continue, -c
Continue indexing on error. (default: False)
--debug, -v, -d, --verbose
Use verbose output. (default: False)
Currently, only files on the file system (--data-type {fs}) are supported.
freva-user-data delete /tmp/user_data/user-freva
freva-databrowser experiment=bias-correct
Results
Status: crawling ...ok
Searching for ESGF data: the freva-esgf command#
The freva-esgf command or the freva esgf sub-command provides
a search syntax to look the model data through all the ESGF portals and
it is derived from
ESGF’s rest API,
although it has been simplified to be used from the command line and
resembles freva-databrowser as close as possible.
Despite the similarities, the two commands rely on different backends
which have different query possibilities.
Note
The Earth System Grid Federation (ESGF) maintains a global system of federated data centers that allow access to the largest archive of model climate data world-wide. ESGF portals are an interface for users to access model data that are distributed in several data centers, also called data nodes, although they themselves do not host any data (e.g. DKRZ). Through them we can access to the output of the climate models contributing to the next assessment report of the Intergovernmental Panel on Climate Change IPCC through the Coupled Model Intercomparison Project CMIP.
Let’s inspect the help menu of the esgf sub-command:
freva-esgf --help
Results
usage: freva esgf [-h] [--datasets] [--show-facet SHOW_FACET] [--opendap]
[--gridftp] [--download-script DOWNLOAD_SCRIPT]
[--query QUERY] [--debug]
[facets ...]
Search/Download ESGF the data catalogue.
positional arguments:
facets Search facet(s). (default: None)
options:
-h, --help show this help message and exit
--datasets List the name of the datasets instead of showing the
urls. (default: False)
--show-facet SHOW_FACET
List all values for the given facet (might be defined
multiple times). The results show the possible values
of the selected facet according to the given
constraints and the number of *datasets* (not files)
that selecting such value as a constraint will result
(faceted search). (default: None)
--opendap Show opendap endpoints instead of http ones. (default:
False)
--gridftp Show gridftp endpoints instead of the http default
ones (or skip them if none found). (default: False)
--download-script DOWNLOAD_SCRIPT
Download wget_script for getting the files instead of
displaying anything (only http). (default: None)
--query QUERY Query fields from ESGF and group them per dataset.
(default: None)
--debug, -v, -d, --verbose
Use verbose output. (default: False)
Similarly to freva-databrowser, freva-esgf expects a list of key=value pairs
in no particular order, but unlike the former it is case sensitive.
For example, given that your Freva instance is configured at DKRZ,
if we want to search the URLs of all the files stored
at the (DKRZ) local node (distrib=false) holding the latest version
(latest=true) of the variable tas (variable=tas) for the
experiment decadal1960 and project CMIP5, then:
freva-esgf project=CMIP5 experiment=decadal1960 variable=tas distrib=false latest=true
Results
http://esgf-node.cmcc.it/thredds/fileServer/esg_dataroot/cmip5/output1/CMCC/CMCC-CM/decadal1960/3hr/atmos/3hr/r1i1p1/v20170725/tas/tas_3hr_CMCC-CM_decadal1960_r1i1p1_196011010000-196012312100.nc
http://esgf-node.cmcc.it/thredds/fileServer/esg_dataroot/cmip5/output1/CMCC/CMCC-CM/decadal1960/3hr/atmos/3hr/r1i1p1/v20170725/tas/tas_3hr_CMCC-CM_decadal1960_r1i1p1_196101010000-196112312100.nc
http://esgf-node.cmcc.it/thredds/fileServer/esg_dataroot/cmip5/output1/CMCC/CMCC-CM/decadal1960/3hr/atmos/3hr/r1i1p1/v20170725/tas/tas_3hr_CMCC-CM_decadal1960_r1i1p1_196201010000-196212312100.nc
http://esgf-node.cmcc.it/thredds/fileServer/esg_dataroot/cmip5/output1/CMCC/CMCC-CM/decadal1960/3hr/atmos/3hr/r1i1p1/v20170725/tas/tas_3hr_CMCC-CM_decadal1960_r1i1p1_196301010000-196312312100.nc
http://esgf-node.cmcc.it/thredds/fileServer/esg_dataroot/cmip5/output1/CMCC/CMCC-CM/decadal1960/3hr/atmos/3hr/r1i1p1/v20170725/tas/tas_3hr_CMCC-CM_decadal1960_r1i1p1_196401010000-196412312100.nc
...
However, be aware that the syntax of the query may change depending
on the targeted dataset, for instance for CMIP6:
freva esgf project=cmip6 product=scenariomip model=cnrm-cm6-1 institute=cnrm-cerfacs experiment=ssp585 time_frequency=3hr variable=uas ensemble=r1i1p1f2 distrib=false latest=true
Results
will not produce any result, while
freva esgf mip_era=CMIP6 activity_id=ScenarioMIP source_id=CNRM-CM6-1 institution_id=CNRM-CERFACS experiment_id=ssp585 frequency=3hr variable=uas variant_label=r1i1p1f2 distrib=false latest=true
Results
http://esgf3.dkrz.de/thredds/fileServer/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_201501010130-202412312230.nc
http://esgf3.dkrz.de/thredds/fileServer/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_202501010130-203412312230.nc
http://esgf3.dkrz.de/thredds/fileServer/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_203501010130-204412312230.nc
http://esgf3.dkrz.de/thredds/fileServer/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_204501010130-205412312230.nc
http://esgf3.dkrz.de/thredds/fileServer/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_205501010130-206412312230.nc
...
will yield the desired output. Note that not only the facets are different from
freva-databrowser but also each value is case sensitive. The naming of the
facets can be, for example, consulted at the corresponding ESGF portal
(e.g. for CMIP6 at DKRZ).
To show the values of certain attributes you can use the --show-facet FACET flag.
For example, for time_frequency :
freva esgf project=CMIP5 experiment=decadal1960 variable=tas distrib=false latest=true --show-facet=time_frequency
Results
[time_frequency]
3hr: 12
day: 19
mon: 20
Similarly to --show-facet, --query QUERY displays key=value metadata pairs (not limited
to attributes only) of selected elements for a search but grouped per dataset.
For example we could query selected metadata information (e.g. mip_era,variable,variant_label,url)
of a certain CMIP6 search and display it grouped by dataset:
freva esgf mip_era=CMIP6 activity_id=ScenarioMIP source_id=CNRM-CM6-1 institution_id=CNRM-CERFACS experiment_id=ssp585 frequency=3hr variable=uas,pr distrib=false latest=true --query=mip_era,variable,variant_label,url
Results
[
{
"mip_era": [
"CMIP6"
],
"url": [
"http://esgf3.dkrz.de/thredds/catalog/esgcet/1586/CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.uas.gr.v20190219.xml#CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.uas.gr.v20190219|application/xml+thredds|THREDDS",
"http://esgf3.dkrz.de/las/getUI.do?catid=649F6BBCDD98B0633CF0042D119CC69D_ns_CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.uas.gr.v20190219|application/las|LAS"
],
"variable": [
"uas"
],
"variant_label": [
"r1i1p1f2"
],
"score": 1.0
},
{
"mip_era": [
"CMIP6"
],
"url": [
"http://esgf3.dkrz.de/thredds/catalog/esgcet/1586/CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.3hr.pr.gr.v20190219.xml#CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.3hr.pr.gr.v20190219|application/xml+thredds|THREDDS",
"http://esgf3.dkrz.de/las/getUI.do?catid=649F6BBCDD98B0633CF0042D119CC69D_ns_CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.3hr.pr.gr.v20190219|application/las|LAS"
],
"variable": [
"pr"
],
"variant_label": [
"r1i1p1f2"
],
"score": 1.0
}
]
To show the name of the datasets instead of the URLs use the --datasets flag:
freva esgf mip_era=CMIP6 activity_id=ScenarioMIP source_id=CNRM-CM6-1 institution_id=CNRM-CERFACS experiment_id=ssp585 frequency=3hr variable=uas variant_label=r1i1p1f2 distrib=false latest=true --datasets
Results
CMIP6.ScenarioMIP.CNRM-CERFACS.CNRM-CM6-1.ssp585.r1i1p1f2.E3hr.uas.gr - version: 20190219
One can retrieve the opendap endpoints instead of the urls as well with --opendap, for example:
freva esgf mip_era=CMIP6 activity_id=ScenarioMIP source_id=CNRM-CM6-1 institution_id=CNRM-CERFACS experiment_id=ssp585 frequency=3hr variable=uas variant_label=r1i1p1f2 distrib=false latest=true --opendap
Results
http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_201501010130-202412312230.nc.html
http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_202501010130-203412312230.nc.html
http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_203501010130-204412312230.nc.html
http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_204501010130-205412312230.nc.html
http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_205501010130-206412312230.nc.html
http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_206501010130-207412312230.nc.html
http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_207501010130-208412312230.nc.html
http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_208501010130-209412312230.nc.html
http://esgf3.dkrz.de/thredds/dodsC/cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_209501010130-210012312230.nc.html
Or the gridftp endpoints instead (--gridftp):
freva esgf mip_era=CMIP6 activity_id=ScenarioMIP source_id=CNRM-CM6-1 institution_id=CNRM-CERFACS experiment_id=ssp585 frequency=3hr variable=uas variant_label=r1i1p1f2 distrib=false latest=true --gridftp
Results
gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_201501010130-202412312230.nc
gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_202501010130-203412312230.nc
gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_203501010130-204412312230.nc
gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_204501010130-205412312230.nc
gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_205501010130-206412312230.nc
gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_206501010130-207412312230.nc
gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_207501010130-208412312230.nc
gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_208501010130-209412312230.nc
gsiftp://esgf3.dkrz.de:2811//cmip6/ScenarioMIP/CNRM-CERFACS/CNRM-CM6-1/ssp585/r1i1p1f2/E3hr/uas/gr/v20190219/uas_E3hr_CNRM-CM6-1_ssp585_r1i1p1f2_gr_209501010130-210012312230.nc
Since the queries yield a list of URLs to the netCDF files
there are not readily available to use. Freva can create a bash script written
around wget to simplify data download with the --download-script /foo/bar.wget
option, where /foo/bar.wget is the destination wget file:
freva esgf --download-script /tmp/script.wget project=CMIP5 experiment=decadal1960 variable=tas distrib=false latest=true
Results
Download script successfully saved to /tmp/script.wget
Note
In order to download the data you will need an ESGF account
(e.g. via DKRZ’s portal),
where after entering a <username> and password
you will get an openID (https://esgf-data.dkrz.de/esgf-idp/openid/<username>)
to use while running the script with the -H flag, e.g.:
> /tmp/script.wget.sh -H
Running script.wget version: 1.3.2
Use script.wget -h for help.
Warning! The total number of files was 2338 but this script will only process 1000.
There were files with the same name which were requested to be download to the same directory. To avoid overwriting the previous downloaded one they were skipped.
Please use the parameter 'download_structure' to set up unique directories for them.
Script created for 916 file(s)
(The count won't match if you manually edit this file!)
Enter your openid : https://esgf-data.dkrz.de/esgf-idp/openid/<username>
Enter password : <password>
Alternatively you can also retrieve the data using certificates, for example through the ESGF PyClient. You will still need your openID for that.
